Introduction to CPPTRAJ
Learning objectives
- Familiarize yourself with AMBER program CPPTRAJ for a more complex
analysis of trajectories.
- Use CPPTRAJ to wrap simulations, calculate RMSDs, RMSFs, and
track angles/bond lengths over trajectory time.
Tutorial
2.1 Introduction to CPPTRAJ
CPPTRAJ is a valuable asset to complement your computational toolkit, as this AMBER program can be used to achieve a more complex analysis of MD trajectories. In this section, you will learn how to use CPPTRAJ to wrap trajectories, provide the distribution of the degree of angle of a trajectory, calculate RMSDs/RMSFs, and find an average structure from a trajectory.
For this tutorial, you will be given multiple CPPTRAJ input files. Each input file contains lines of commands for CPPTRAJ, but they can each be individually submitted in the interactive command line of CPPTRAJ. The interactive command line of CPPTRAJ can be accessed by typing into your terminal:
[user@expanse] cpptraj
Which will open up the interactive command line seen below:
[user@expanse] cpptraj
CPPTRAJ: Trajectory Analysis. V4.26.4 (AmberTools V20.00)
___ ___ ___ ___
| \/ | \/ | \/ |
_|_/\_|_/\_|_/\_|_
| Date/time: 00/00/24 00:00:00
| Available memory: 54.268 GB
To get out of CPPTRAJ, type quit.
2.2 Wrapping Trajectories
In this section, you will explore acetic acid in solution and acid acid in the gas phase. Your goal is to understand how the dihedral angle behaves in each environment. However, before you start the analysis on the trajectories of each system, you must first wrap the systems. Wrapping a trajectory means adjusting the positions of the molecules in a simulation so that they can stay within your defined simulation box. Without wrapping a trajectory, visually the molecules within the system can appear to look separated, drifting away or blown out due to the periodic boundary conditions. Therefore, it can be hard to gain an accurate interpretation of what is true behavior displayed in your system. If you are wanting to visually inspect your system, wrapping should be the first step prior to trajectory analysis to avoid confusion. Wrapping is one of CPPTRAJ’s most essential roles.
CPPTRAJ can wrap both restart files and trajectory files. In this case, you will be wrapping the trajectory file of acetic acid in solution. First, you must decide on a center in which to wrap everything around. Usually, this is the ligand. Therefore, in this tutorial, it will be ligand L01 (acetic acid). Let’s start with the acetic acid in the gas phase trajectory first.
Find the directory containing the acetic acid input files: /expanse/projects/qstore/amber_ws/tutorials/VMD_CPPTRAJ_Basics/input_files/acetic_acid.
Access the CPPTRAJ interactive command line:
[user@expanse] cpptraj
Similarly to VMD, you must load the topology file first, followed by the trajectory file. CPPTRAJ’s command to load a topology file is parm.
Type the following into your command line:
> parm acetic_acid_aq.parm7
Then, load the trajectory file with the trajin command. WindowsUsers: load DCD trajectory file.
Type the following:
> trajin acetic_acid_aq.nc
Next, the command reference is used to tell CPPTRAJ to use a specified set of coordinates to be the reference structure. It is good practice for the reference structure to be wrapped and centered itself.
Type this:
> reference centered.acetic_acid_aq.rst7
Then, use the command center to center the specified atoms in the atom mask.
> center :L01 mass
The atom selection is the atom(s)/molecule that we are wanting to be the center. The mask :L01 specifies all the atoms in residue L01. mass indicates that the centering should be done based on the center of mass of the atom selection. Lastly, reference tells cpptraj that the centering should be related to the reference structure that has previously been loaded.
Type:
> autoimage :L01
Type:
> center :L01 reference
Use the command trajout and specify the filename of the newly wrapped trajectory output product in either NetCDF or DCD format.
> trajout centered.acetic_acid_aq.nc
Once you have entered all of that:
Type run or go to execute the job.
> run
Once the job is finished running, type quit to quit CPPTRAJ and the interactive command line.
> quit
Your interactive terminal of CPPTRAJ should look like this:
[user@expanse] cpptraj
> parm acetic_acid_aq.parm7
[parm acetic_acid_aq.parm7]
Reading 'acetic_acid_aq.parm7' as Amber Topology
Radius Set: modified Bondi radii (mbondi)
> trajin acetic_acid_aq.nc
[trajin acetic_acid_aq.nc]
Reading 'acetic_acid_aq.nc' as Amber NetCDF
> reference centered.acetic_acid_aq.rst7
[reference centered.acetic_acid_aq.rst7]
Reading 'centered.acetic_acid_aq.rst7' as Amber Restart
Setting active reference for distance-based masks: 'centered.acetic_acid_aq.rst7'
> center :L01 mass
[center :L01 mass]
CENTER: Centering coordinates using center of mass of atoms in mask (:L01) to box center.
> autoimage :L01
[autoimage :L01]
AUTOIMAGE: To box center based on center of mass, anchor mask is [:L01]
> center :L01 mass reference
[center :L01 mass reference]
CENTER: Centering coordinates using center of mass of atoms in mask (:L01) to center of mask (:L01) in reference 'centered.acetic_acid_aq.rst7'.
> trajout centered.acetic_acid_aq.nc
[trajout centered.acetic_acid_aq.nc]
Writing 'centered.acetic_acid_aq.nc' as Amber NetCDF
> run
[run]
---------- RUN BEGIN -------------------------------------------------
PARAMETER FILES (1 total):
0: acetic_acid_aq.parm7, 11740 atoms, 2946 res, box: Orthogonal, 2946 mol, 2929 solvent
INPUT TRAJECTORIES (1 total):
0: 'acetic_acid.nc' is a NetCDF AMBER trajectory with coordinates, velocities, time, box, Parm acetic_acid_aq.parm7 (Orthogonal box) (reading 21 of 21)
Coordinate processing will occur on 21 frames.
REFERENCE FRAMES (1 total):
0: centered.acetic_acid_aq.rst7:1
Active reference frame for distance-based masks is 'Cpptraj Generated Restart'
OUTPUT TRAJECTORIES (1 total):
'centered.acetic_acid_aq.nc' (21 frames) is a NetCDF AMBER trajectory
BEGIN TRAJECTORY PROCESSING:
.....................................................
ACTION SETUP FOR PARM 'acetic_acid_aq.parm7' (3 actions):
0: [center :L01 mass]
Mask [:L01] corresponds to 8 atoms.
1: [autoimage :L01]
Anchoring on atoms selected by mask ':L01'
Mask [:L01] corresponds to 8 atoms.
Mask [:L01] corresponds to molecule 1
2945 molecules are mobile.
2: [center :L01 mass reference]
Mask [:L01] corresponds to 8 atoms.
.....................................................
ACTIVE OUTPUT TRAJECTORIES (1):
centered.acetic_acid_aq.nc (coordinates, velocities, time, box)
----- acetic_acid_aq.nc (1-21, 1) -----
0% 10% 20% 30% 40% 50% 60% 70% 80% 90% 100% Complete.
Read 21 frames and processed 21 frames.
TIME: Avg. throughput= 202.0474 frames / second.
ACTION OUTPUT:
TIME: Analyses took 0.0000 seconds.
RUN TIMING:
TIME: Init : 0.0001 s ( 0.14%)
TIME: Trajectory Process : 0.1039 s ( 98.88%)
TIME: Action Post : 0.0000 s ( 0.00%)
TIME: Analysis : 0.0000 s ( 0.00%)
TIME: Data File Write : 0.0000 s ( 0.00%)
TIME: Other : 0.0010 s ( 0.01%)
TIME: Run Total 0.1051 s
---------- RUN END ---------------------------------------------------
> quit
[quit]
--------------------------------------------------------------------------------
To cite CPPTRAJ use:
Daniel R. Roe and Thomas E. Cheatham, III, "PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data". J. Chem. Theory Comput., 2013, 9 (7), pp 3084-3095.
The wrapped trajectory is now called centered.acetic_acid_aq.nc.
To submit the input file to cpptraj, type this into your command line:
[user@expanse] cpptraj -i ctraj.wrap_traj_acetic_acid_aq.in
Windows Users: As previously mentioned, Windows does not support the NetCDF plugin for VMD, and the proper DCD trajectory file was provided for you. Here, you can practice converting NetCDF files to DCD files yourself by running the bash script nc2dcd.sh. This file creates ctraj.convert_nc_dcd.in and submits it to CPPTRAJ. The file contains:
#! /bin/bash
# Define the script name
thisScript='basename "$0"'
# Check if the number of arguments is exactly 2
if ! [ "$#" -eq 2 ]; then
cat <<EOF
Needs two file names:
$thisScript [parm].parm7 [traj].nc
EOF
exit 0
fi
# Load the appropriate module files
module purge
module load workshop/amber24
# Define arguments
parm_file=$1
traj_file=$2
# Remove the .nc extension
if [[ $traj_file == *.nc ]]; then
traj_base=${traj_file%.nc}
else
traj_base=$traj_file
fi
# Create CPPTRAJ input file
cat <<EOF > ctraj.convert_nc_dcd.in
parm ${parm_file}
trajin ${traj_base}.nc
trajout ${traj_base}.dcd
run
quit
EOF
# Submit the CPPTRAJ input file
cpptraj -i ctraj.convert_nc_dcd.in > c.log
rm c.log
This script needs to be fed two arguments to run: the first is the topology file and the second is NetCDF trajectory file.
To convert centered.acetic_acid_vac.nc to centered.acetic_acid_vac.dcd, submit the bash script:
[user@expanse] bash nc2dcd.sh acetic_acid_vac.parm7 centered.acetic_acid_vac.nc
You should now see in your directory centered.acetic_acid_vac.dcd and the CPPTRAJ input file, ctraj.convert_nc_dcd.in, that was created and submitted.
2.3 Trajectory Time Series and Distribution Analysis
Early on in this tutorial, you measured a dihedral angle of a static structure of acetic acid in solution using VMD. You will be examining the same dihedral angle of the acetic acid molecule when it is in the gas phase and when it is in solution. The aim of this section is to track the movement of the hydrogen in the hydroxyl group throughout each of the trajectories. Furthermore, you will be introduced to xmgrace in order to create a graph comparing the degree of the dihedral angle in different environments.
The xmgrace module on Expanse conflicts with AMBER on Expanse. Therefore, it is recommended to open another Expanse window, To load the xmgrace module, you will need to load:
[user@expanse] module load cpu/0.15.4 intel/19.1.1.217 grace/5.1.25
Then load the xmgrace module:
[user@expanse] module load grace
To access xmgrace, use:
[user@expanse] xmgrace
The script that you will submit CPPTRAJ for acetic acid in gas is called ctraj.dihedral_vac.in and looks like this:
parm acetic_acid_vac.parm7
trajin centered.acetic_acid_vac.nc
dihedral :L01@H4 :L01@O2 :L01@C2 :L01@C1 out dihedral_acetic_acid_vac.dat
run
quit
The submit script for acetic acid in solution, ctraj.dihedral_aq.in, is:
parm acetic_acid_aq.parm7
trajin centered.acetic_acid_aq.nc
dihedral :L01@H4 :L01@O2 :L01@C2 :L01@C1 out dihedral_acetic_acid_aq.dat
run
quit
Submit each input file one at a time, via the command line in your terminal:
[user@expanse] cpptraj -i ctraj.dihedral_vac.in
[user@expanse] cpptraj -i ctraj.dihedral_aq.in
Look for the two files dihedral_acetic_acid_vac.dat and dihedral_acetic_acid_aq.dat. You will use a common graphing program called xmgrace to open these files.
To do so, type:
[user@expanse] xmgrace dihedral_acetic_acid_vac.dat dihedral_acetic_acid_aq.dat
Which will bring up the following:
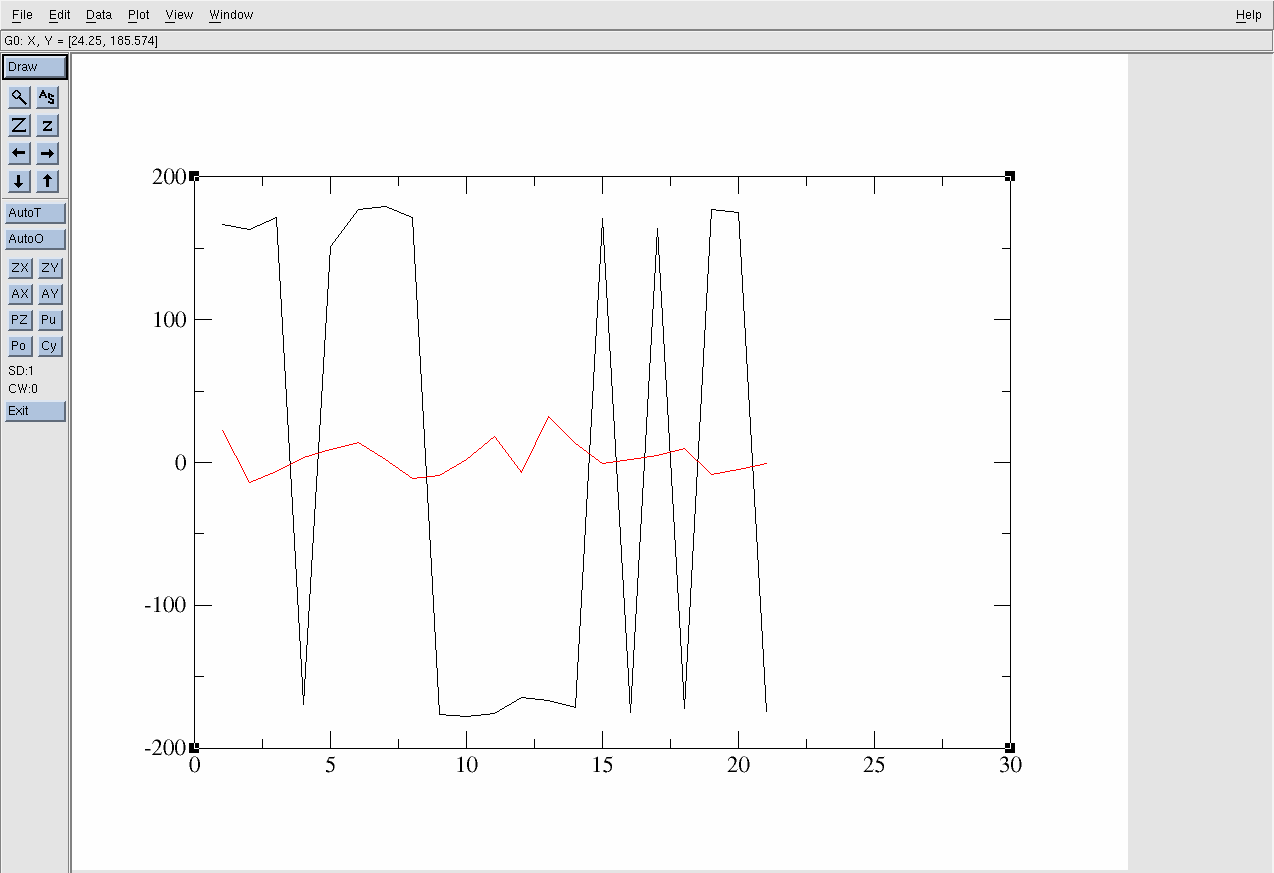
Figure 18. xmgrace interface.
The black line is the acetic acid molecule in the gas phase and the red line is it in the solution environment. The x-axis is the Frame and the y-axis is the degree of the Dihedral angle.
However, the current range of the y-axis (-200 to 200) hinders an intuitive understanding of the dihedral angle fluctuation. Therefore, it is necessary to transform the data to be in the range of -90° to +270°. To achieve this:
Go to Data→Transformations→Evaluate Expression.
Figure 19. Evaluate Expression panel.
Conveniently in xmgrace, you can easily manipulate your data. To transform the y-values to be within the range -90° to +270°:
Find and select G0.S0[2][21]. (This is the acetic acid in the gas phase data set and will be highlighted in black when selected).
In the Formula section, type the following: y=(y+360) % 360.
Press Apply.
This will produce a new data set called G0.S2[2][21] graphed in a green line:
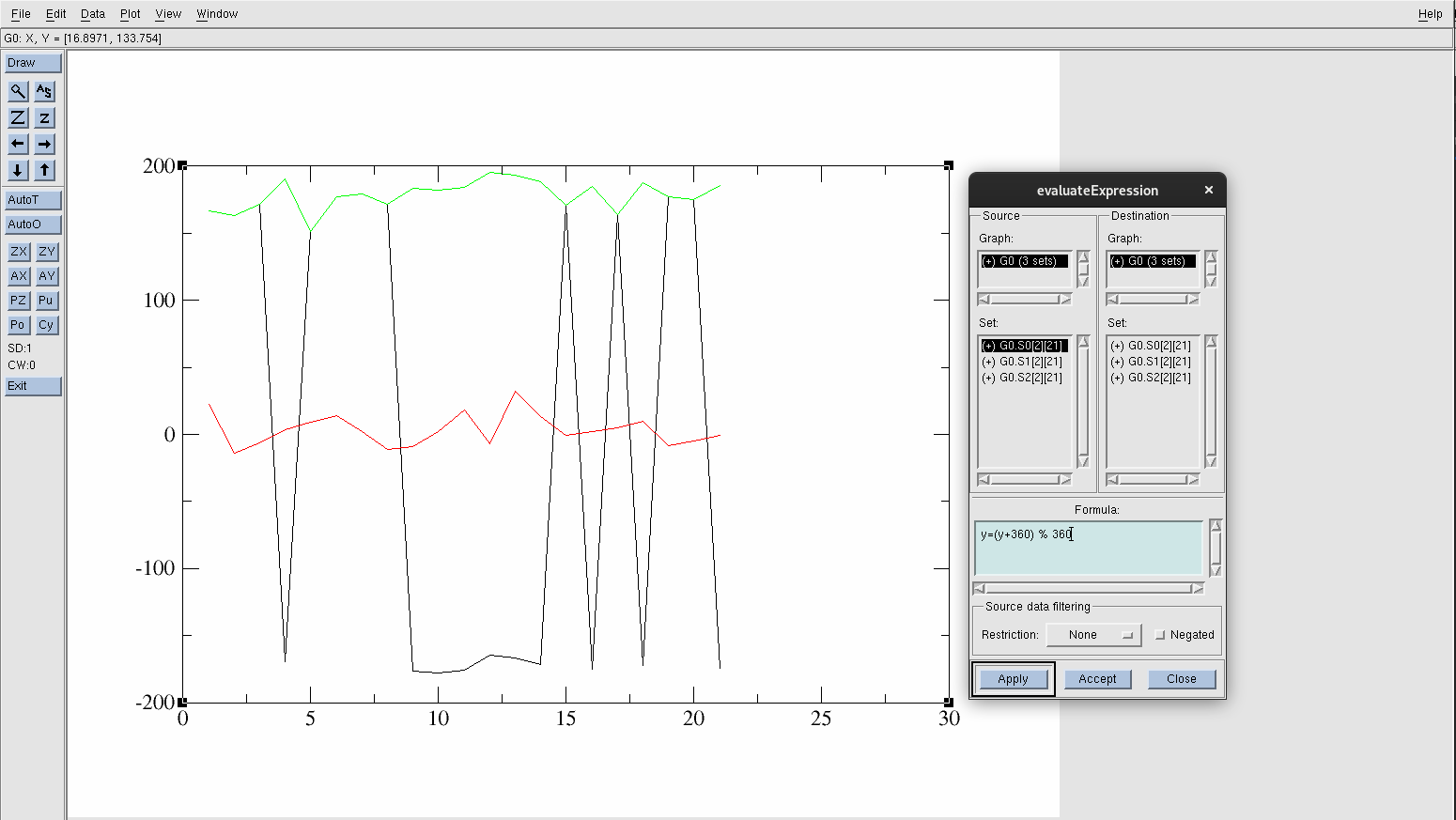
Figure 20. Shifted y-values.
This formula shifts the y-values by 360° to bring all the negative angles into a positive range. The % 360 operation ensures that the new values will be within the 0 to 360 range. If any of the angles exceed 360°, they are wrapped. For example, 370° would become 10°.
Hide the original data set by going to Plot→Set Appearance.
Find and select G0.S0[2][21]. Right-click and choose Hide. You know it will be hidden if the (+) in front of the data set changes to (-).
Select G0.S1[2][21] and increase the width under Line Properties to 2.5.
Hit Apply.
Select G0.S2[2][21] and increase the width under Line Properties to 2.5.
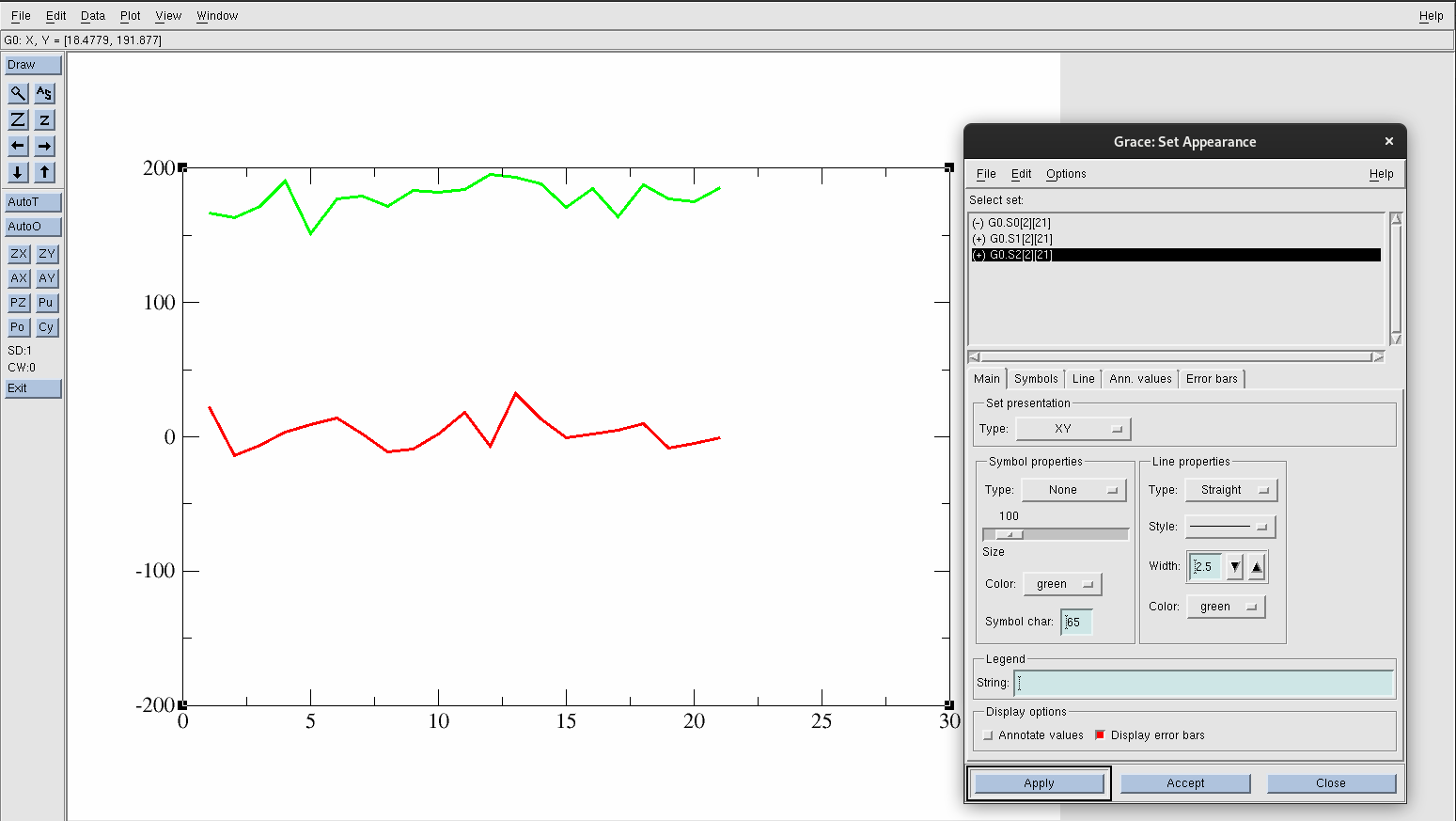
Figure 21. Isolated data.
The AS button on the left-side panel of Xmgrace will always adjust the axis automatically for you.
Press the AS button. You should now only see a green line (acid acid in gas with transformed y-values) and a red line (acetic acid in solution)
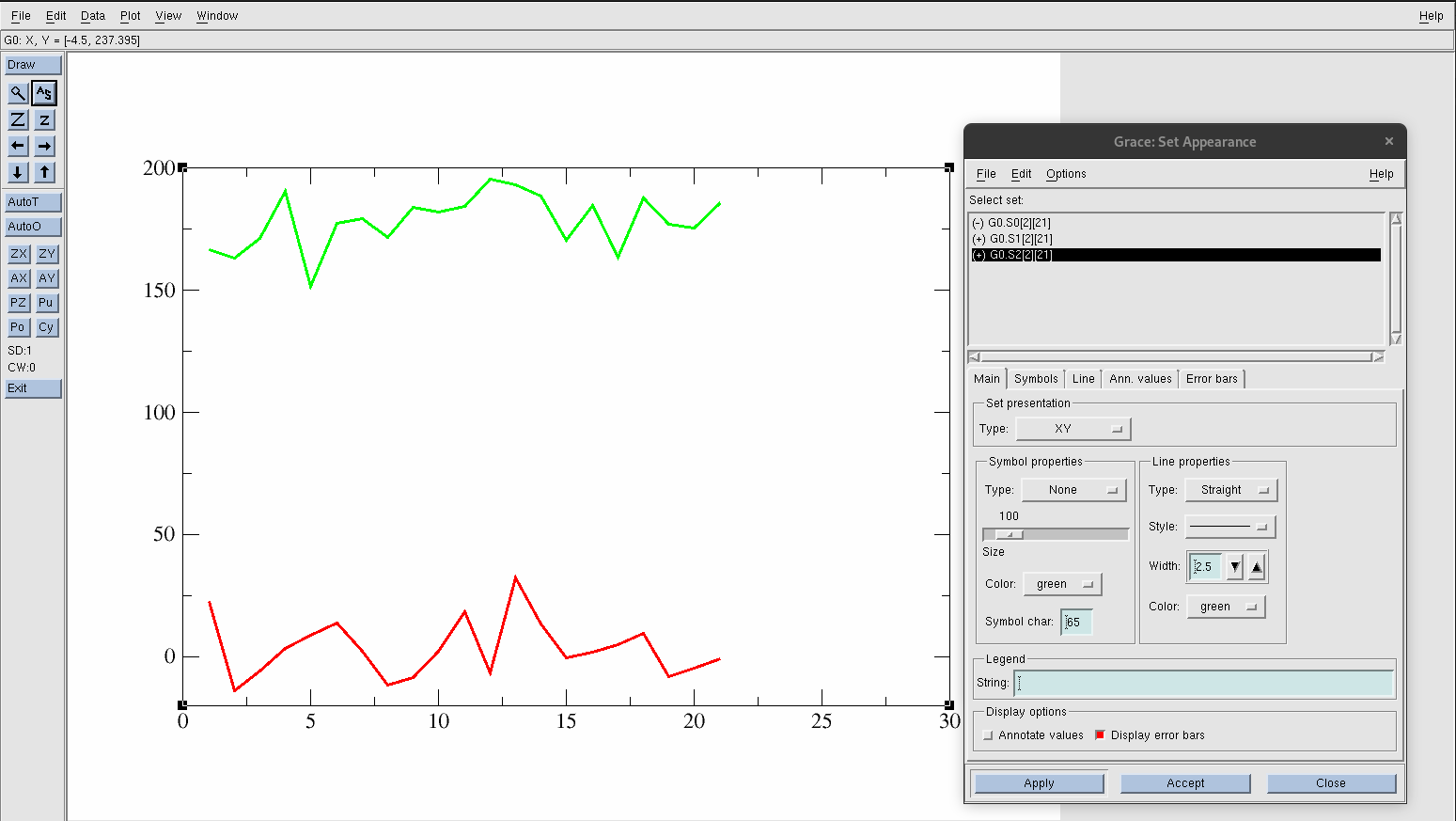
Figure 22. Readjusted graph after clicking the autoscale button.
The x-axis is the number of frames, while the y-axis is the dihedral angle in °. To change the range of the axes:
Go to Plot→Axis Properties.
The Edit: should read X axis.
Find the Start and Stop boxes.
Change the Start to 1.
Change the Stop to 21.
Press Apply.
To label the axes:
Find Label String.
Type Frame.
Press Apply.
To change the axis label and values of the y-axis:
Select Y axis from Edit:.
Find the Start and Stop boxes.
Change the Start to -90.
Change the Stop to 270.
Press Apply.
To label the axes:
Find Label String.
Type Dihedral (\c0\C).
Press Apply.
Save this graph by:
Go to File→Save As.
Save the file as acetic_acid_frameVSdihedral.agr
To save the graph as a PNG file type:
Go to File→Print Setup.
Under Device setup, change the PostScript to PNG.
Then, under Page, find the Dimensions line.
Click pix, and change to in.
Change the Resolution to 300.
Hit Apply.
Go back to File and click on Print.
Your graph should look like:
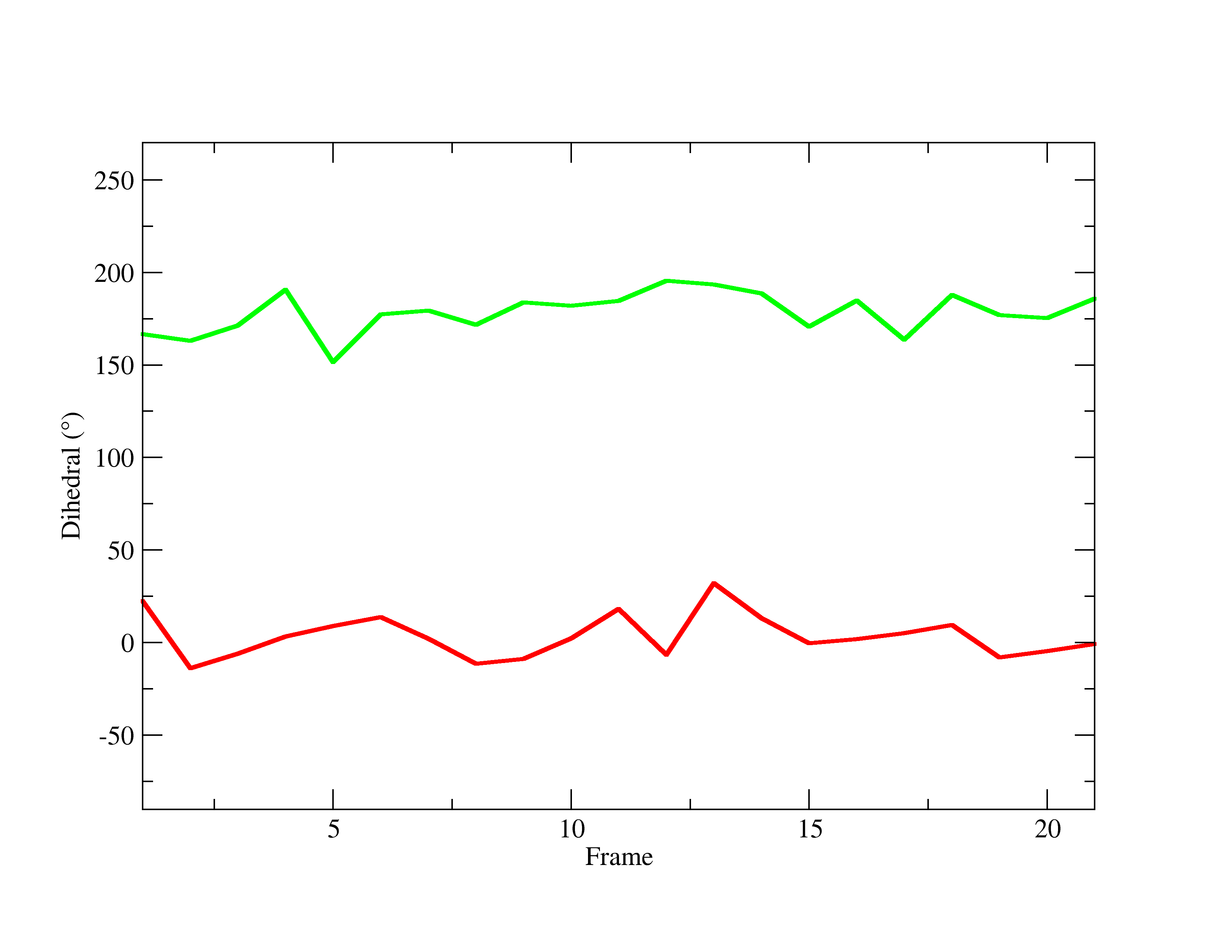
Figure 23. Final PNG of trajectory frame number vs. Acetic Acid’s dihedral angle in both the gas phase(green) and in solution (red) .
As you can see from the graph, the H4 in the hydroxyl group behaves differently in each environment, fluctuating within a different angle range. To obtain a more meaningful analysis, create a histogram to see the distribution of the data. To do so:
Go to Data→ Transformations→ Histograms.
The window will pop up. Create a histogram for acetic acid in solution first.
Select G0.S1[2][21].
Into Start at: type -15.
Into Stop at: type 35.
Change the # of bins: to 5
Hit Apply.
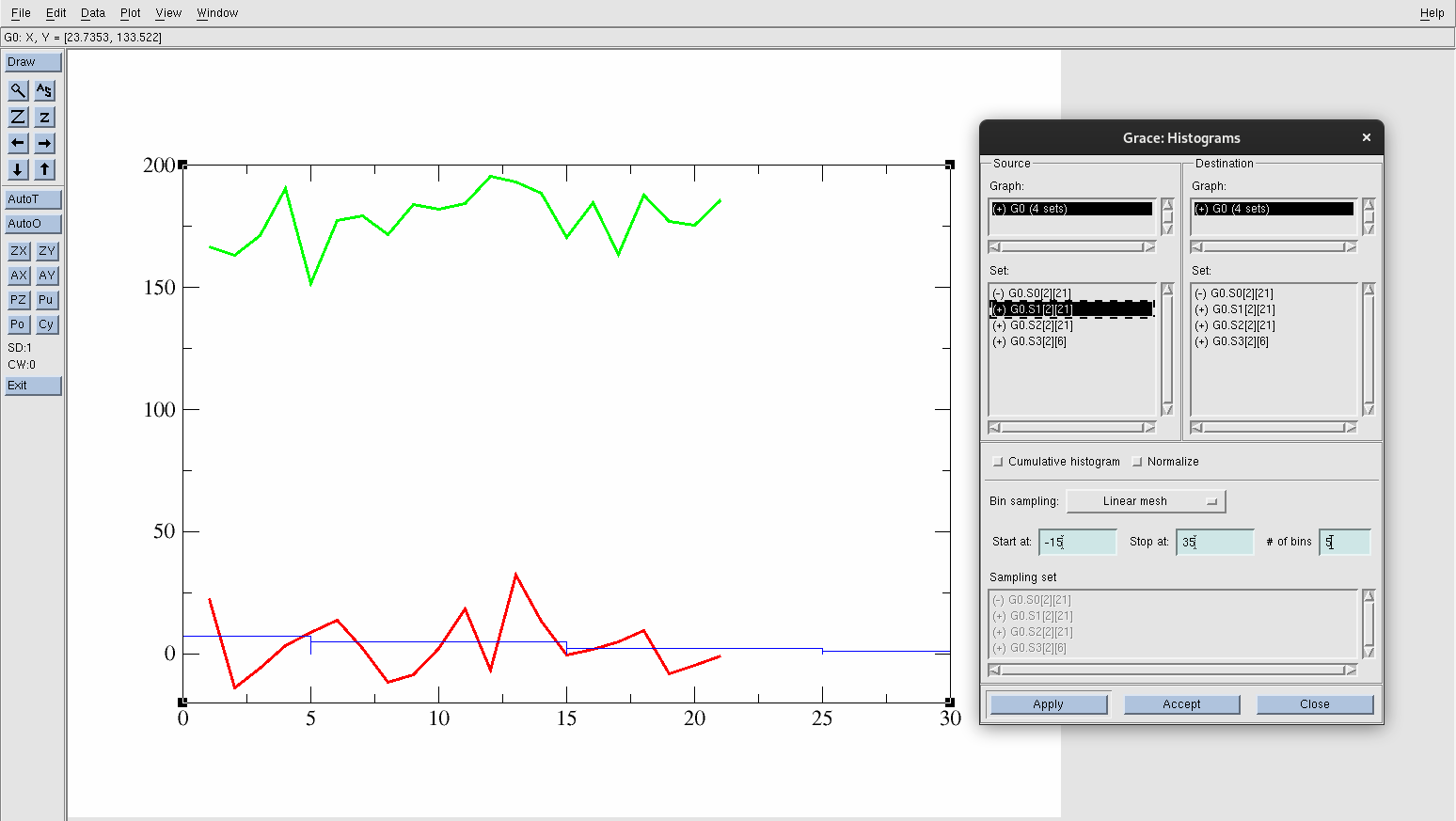
Figure 24. Created histogram for acetic acid in solution data.
Select G0.S2[2][21].
Into Start at: type 150.
Into Stop at: type 197.
Change the # of bins: to 5
Hit Apply.
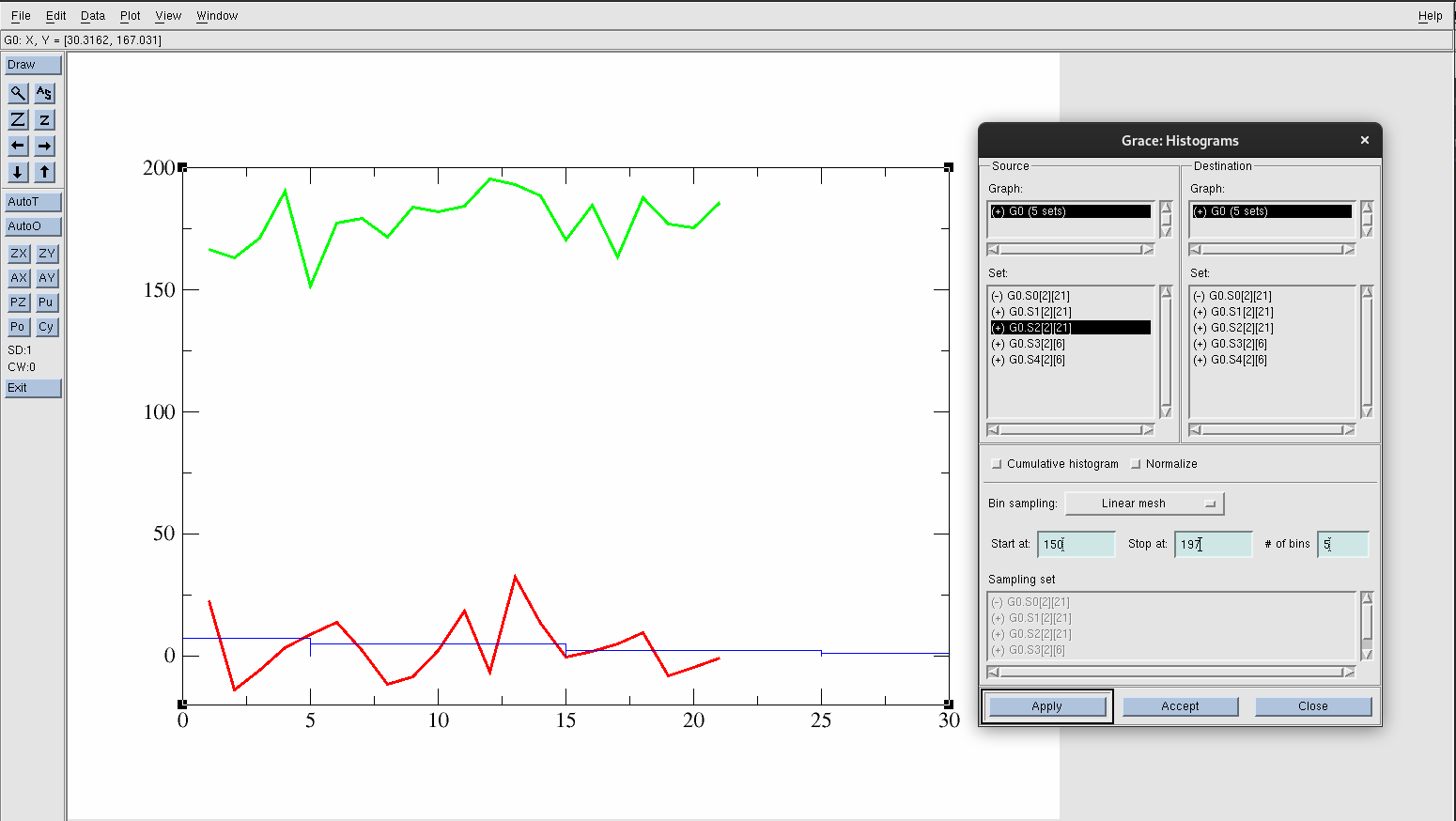
Figure 25. Created histogram for acetic acid in gas data.
To see the histograms made:
Go back to Plot→Set Appearance.
Hide data set G0.S1[2][21] and G0.S2[2][21].
Press the AS button again.
Select G0.S3[2][21], and change the color to red.
Change the width of the line to 2.5.
Select G0.S4[2][21], and change the color to green.
Change the line width of G0.S4[2][21] to 2.5.
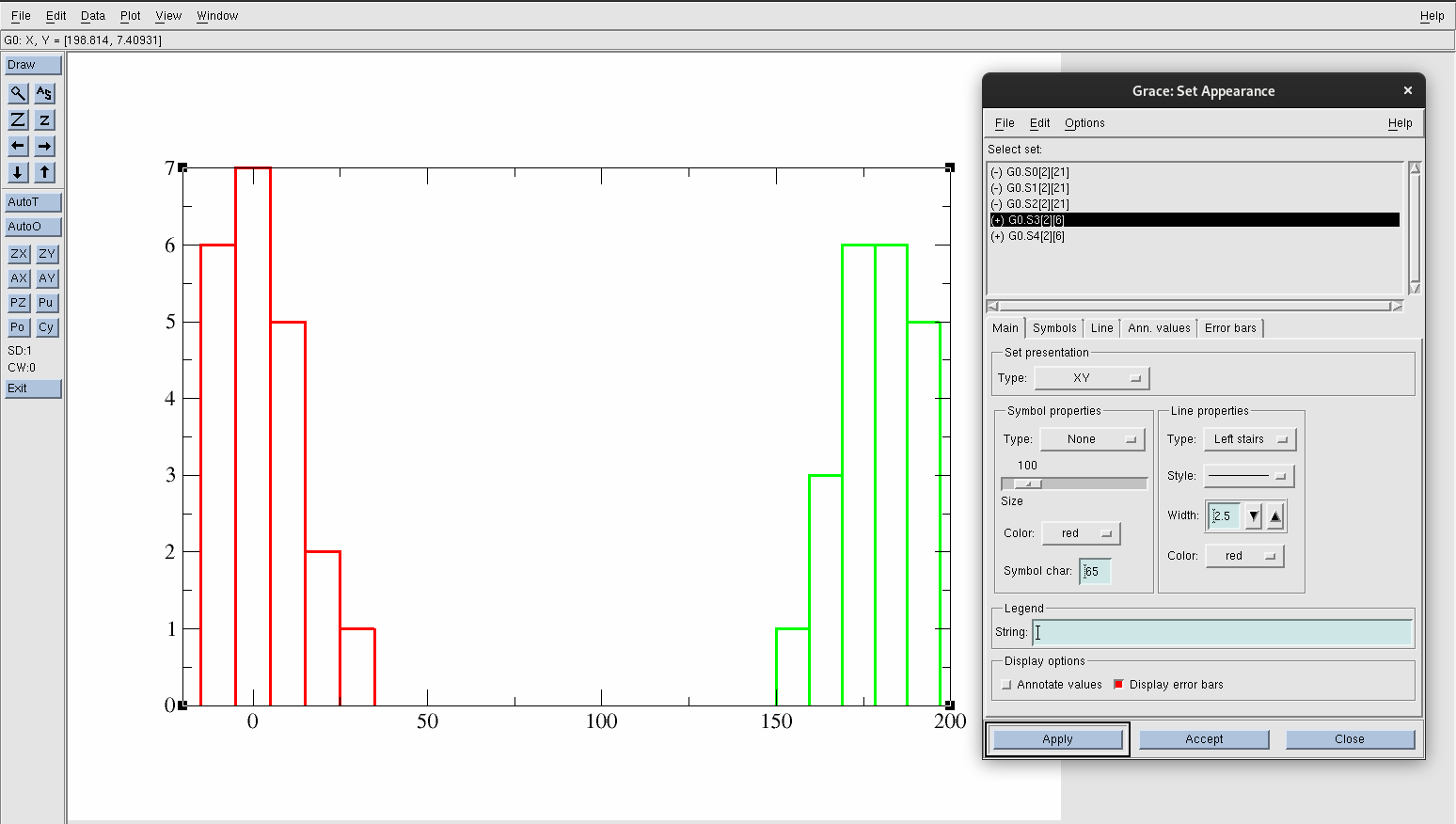
Figure 26. Distributions of the dihedral angle of acetic acid in both environments.
Now, change the axes by again going to:
Plot→Axis Properties.
The Edit: should read X axis.
Find the Start and Stop boxes.
Change the Start to -90.
Change the Stop to 270.
Press Apply.
To label the axes:
Find Label String.
Type Dihedral (\c0\C).
Press Apply.
To change the axis label and values of the y-axis:
Select Y axis from Edit:.
Find the Start and Stop boxes.
Change the Start to 0.
Change the Stop to 10.
Press Apply.
To label the axes:
Find Label String.
Type Frequency.
Press Apply.
Save this graph by:
Go to File→Save As.
Save the file as acetic_acid_dihedral_distru.agr.
To save the graph as a PNG file type:
Go to File→Print Setup.
Under Device setup, change the PostScript to PNG.
Change the file name to match the *.agr file name: acetic_acid_dihedral_distru.png.
Then, under Page, find the Dimensions line.
Click pix, and change to in.
Change the Resolution to 300.
Hit Apply.
Go back to File and click onPrint.
Your graph should look like:
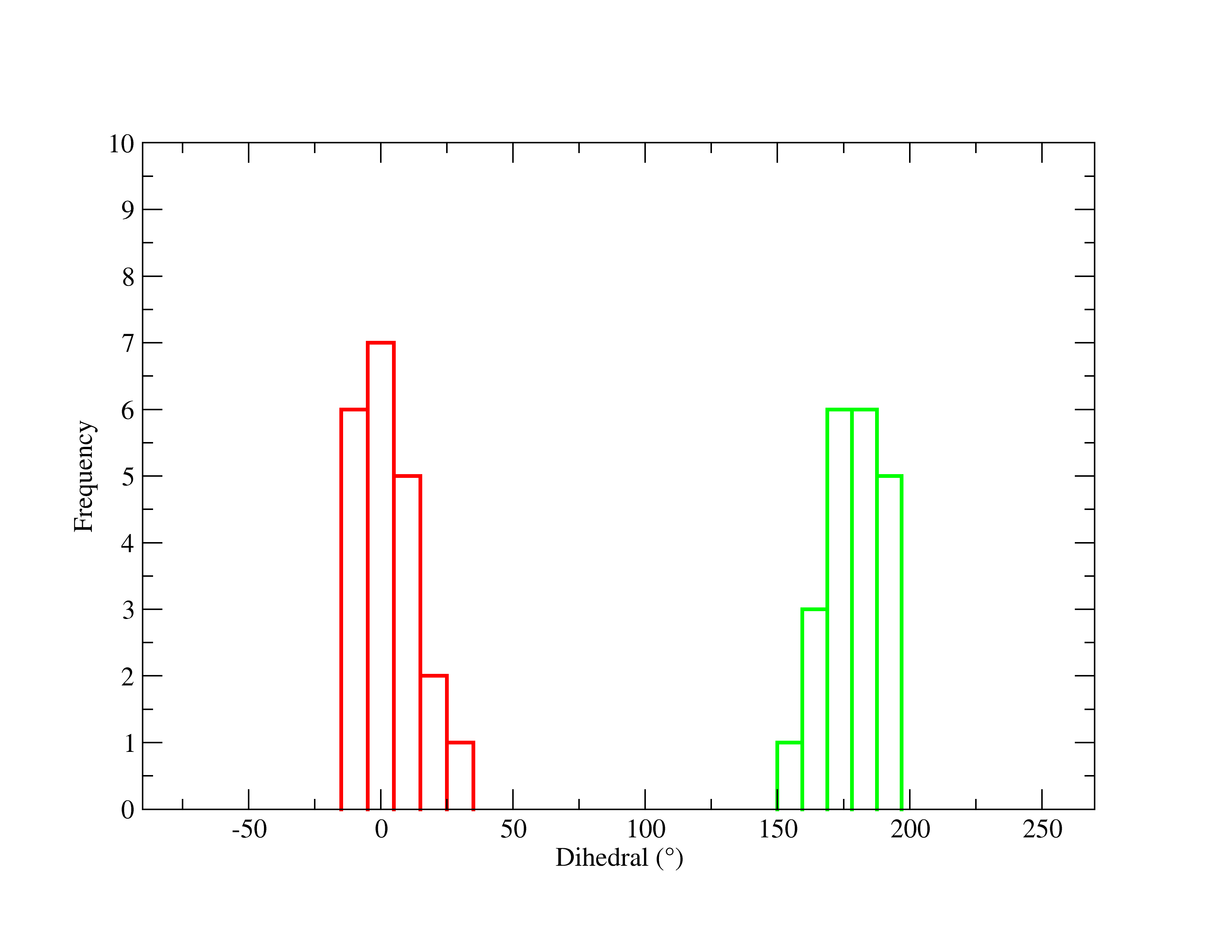
Figure 27. Final PNG image of the distributions of the dihedral angle of acetic acid in both environments.
This display is now the distribution graph of the dihedral angles in each system, with the x-agis now as the Dihedral in degrees and the y-axis representing the frequency. The green line is acetic acid in gas and the red line is acetic acid in solution.
Return back to VMD to visually inspect the changes occuring throughout the simulation of the molecule in each system. Visualizing structures is a crucial aspect for MD simulations, as it helps to interpret your numerical data. Furthermore, it is a good practice to constantly look at your structure to ensure that it remains physically realistic and retains your desired properties.
To pull up each of these trajectories in VMD:
Type the following into your teminal, remember, Windows Users will need to load DCD trajectory files:
[user@expanse] vmd -f acetic_acid_vac.parm7 centered.acetic_acid_vac.nc -f acetic_acid_aq.parm7 centered.acetic_acid_aq.nc
This will load each system under one VMD window. The -f flag loads all subsequent files into the same molecule. Meaning, we loaded both systems by first loading the corresponding topology and trajectory file for acetic acid in the gas phase, then the files for acetic acid in solution. Once in VMD, go to Extensions→Analysis→RMSD Trajectory Tool, type resname L01 and then press Align. In your Representations create resname L01 with CPK as the Drawing Method. You should see the acetic acid molecules from each system overlayed on top of each other:
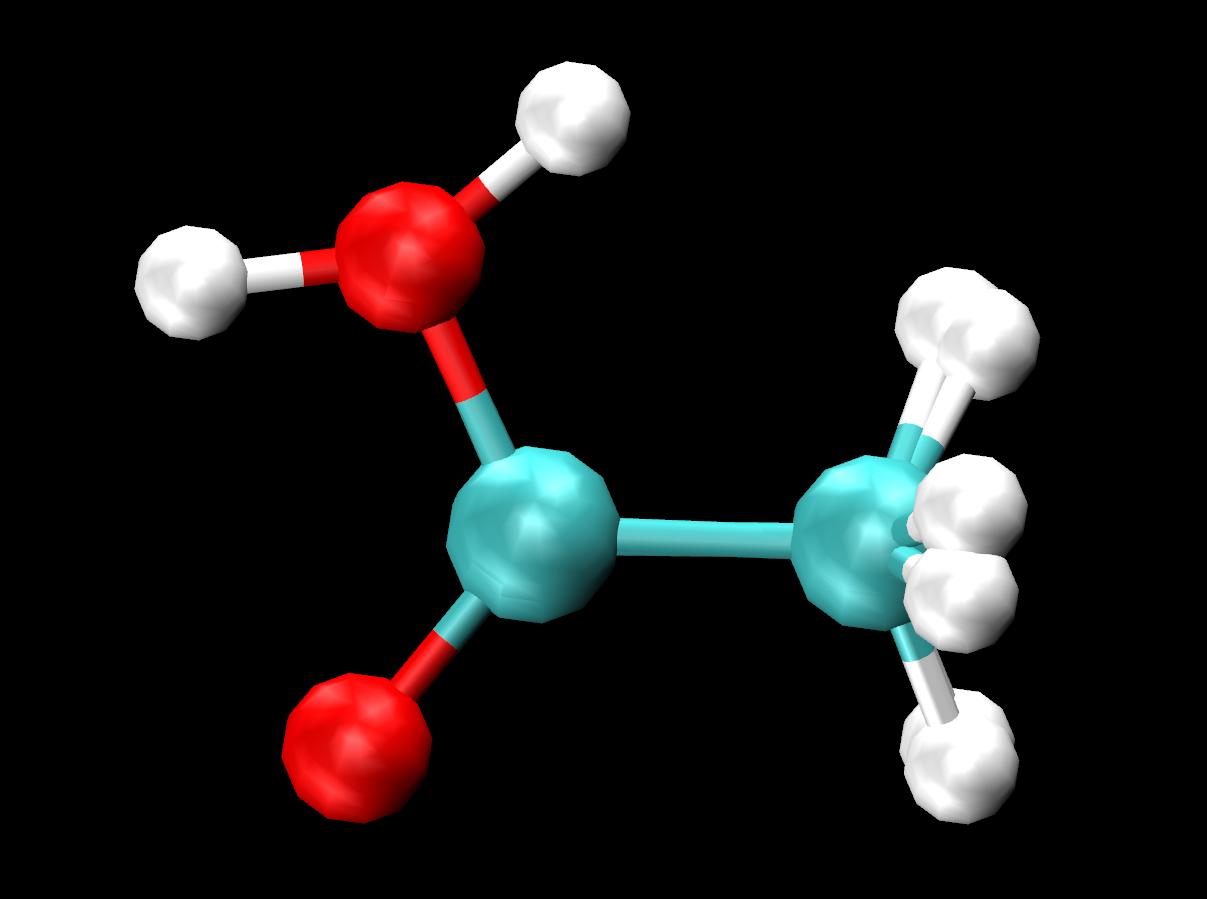
Figure 28. Overlay of acetic acid molecule from each trajectory.
You can turn off one representation at a time to clearly see the difference in orientation of the hydrogen (H4) within the hydroxyl group. You can turn off the representations or in the VMD Main, turn off the display of the acetic acid in solution system completely, (by clicking D) The H4 looks to be making an intramolecular bond with O1 (within the C=O) in the gas phase, but does not show the same in solution. What could be the reasoning behind this change in behavior?
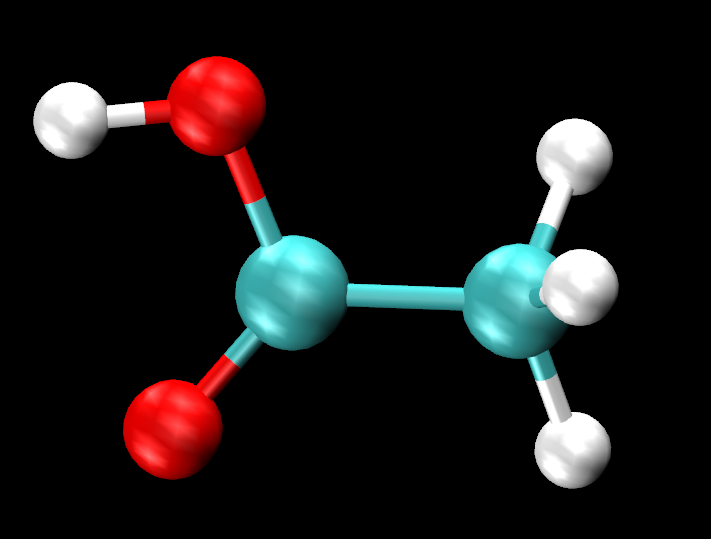
Figure 29. Acetic acid molecule in gas phase.
Figure 30. Acetic acid molecule in solution.
2.4 Computing Average Structures and Fluctuations
You have already learned how to create RMSD plots using VMD. With the same system, MTR1, you will now use CPPTRAJ to create RMSDs. Additionally, you will learn how to calculate RMSFs and find the average structure of simulations using CPPTRAJ.
Go to the directory that contains the MTR1 files: /expanse/projects/qstore/amber_ws/tutorials/VMD_CPPTRAJ_Basics/input_files/acetic_acid.
You will find a script called ctraj.RMSD_MTR1.in. The contents of this script is:
parm MTR1_prot-run.parm7
trajin centered.MTR1_prot-run.nc
rms ToFirst :1-69&!@H= first out RMSD_MTR1.dat mass
run
quit
This input file again follows a standard of inputting your topology file first, then your trajectory. The rms ToFirst :1-69&!@H= first out RMSD_MTR1.dat mass line tells cpptraj to do an RMSD calculation, saving the data set as “ToFirst”, using all non-hydrogen atoms in the residues 1 through 69. This residue selection is the entire nucleic molecule. The first is telling cpptraj to use the first frame in the trajectory as a reference, with out as the command to write the output to a file named RMSD_MTR1.dat. The mass command indicates a mass-weighted RMSD calculation.
Submit this file as your did before:
[user@expanse] cpptraj -i ctraj.RMSD_MTR1.in
Once the job is complete, the RMSD_MTR1.dat file will be in your directory.
Using xmgrace, open this file:
[user@expanse] xmgrace RMSD_MTR1.dat
Which will pop up:
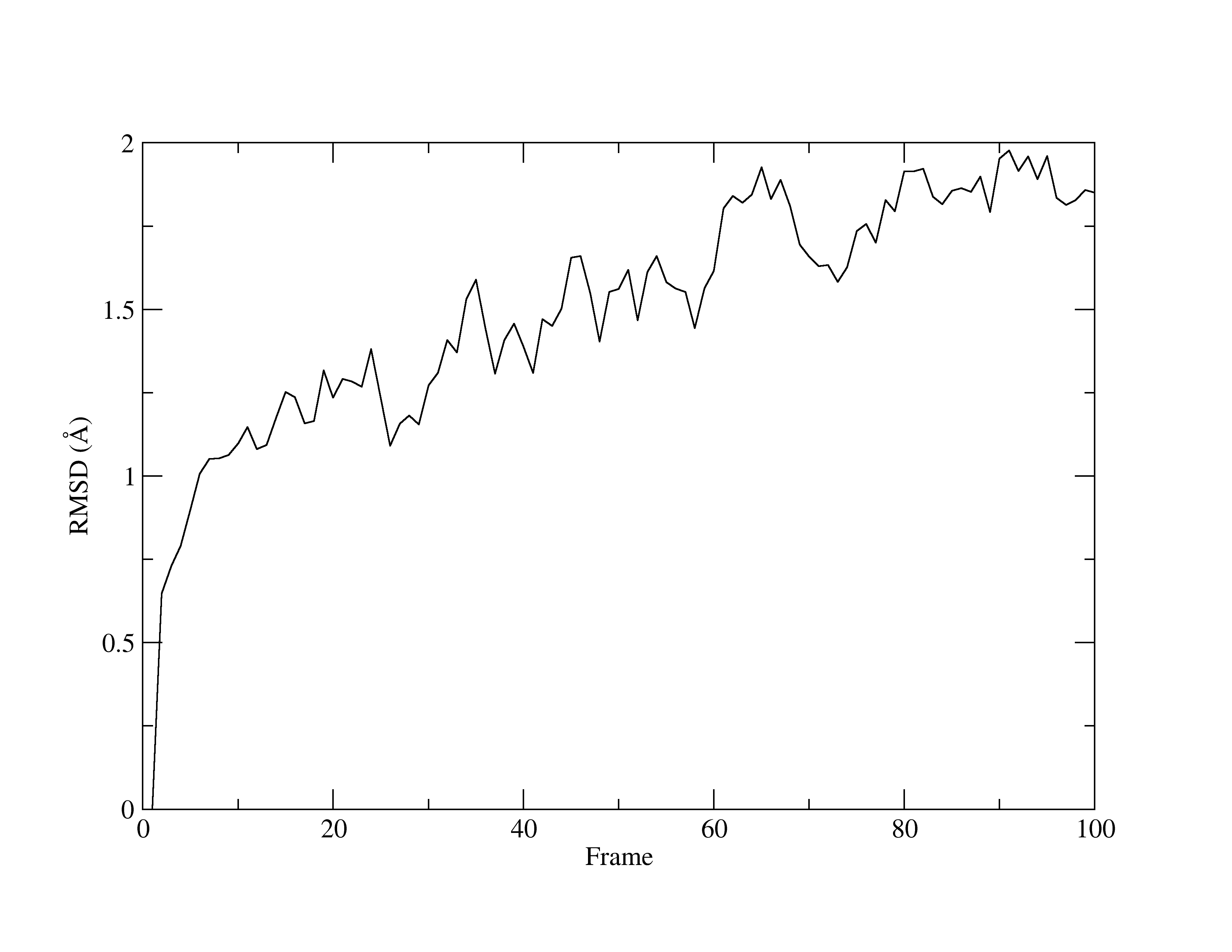
Figure 31. RMSD of MTR1.
With a quick glance of the graph, you will see that it is comparable to the same graph that was produced by VMD. Once again, the line fluctuates and has an upwards direction. Yet, given the very low values for RMSD, you can concur that the structure is stable.
Next, you will learn to use CPPTRAJ to calculate a Root Mean Square Fluctuation (RMSF). Like RMSDs, RMSFs are important analytical measurements that provide insight into your system. RMSFs quantify the overall flexibility and mobility of each atom or residue within your system as the simulation progresses. RMSFs calculate how much the position of each atom in your selection deviates from its average position throughout the simulation time.
In your directory, find the input file, ctraj.RMSF_MTR1.in.
Open in VIM to see the script:
parm MTR1_prot-run.parm7
reference centered.MTR1_prot-run.rst7
trajin centered.MTR1_prot-run.nc
rms :1-69&!@H reference mass
atomicfluct out RMSF_MTR1.dat :1-69&!@H= byres
atomicfluct out RMSF_Bfact_MTR1.dat :1-69&!@H= byres bfactor
run
quit
The script is similar to the previous one, but there are two new lines containing the RMSF commands.atomicfluct out RMSF.dat :1-69&!@H= byres and atomicfluct out RMSF_Bfact.dat :1-69&!@H= byres bfactor. atomicfluct is the command for an RMSF calculation. As seen before, out directs CPPTRAJ to output the data into a file name of your choosing (here, it is RMSF.dat, followed by your atom selection. The keyword byres specifies the output to be per residue, which sums the fluctuations of all atoms in each residue. The other keyword seen, bfactor, which will calculate the B-factor of the MD trajectory. This is an additional analysis one can do to compare simulation to crystallographic data. A low B-factor is indicative of a well-ordered and stable structure, while a high b-factor suggests more movement and thereby, greater fluctuations.
Open the RMSF.dat by using xmgrace:
[user@expanse] xmgrace RMSF_MTR1.dat
Which will look like this:
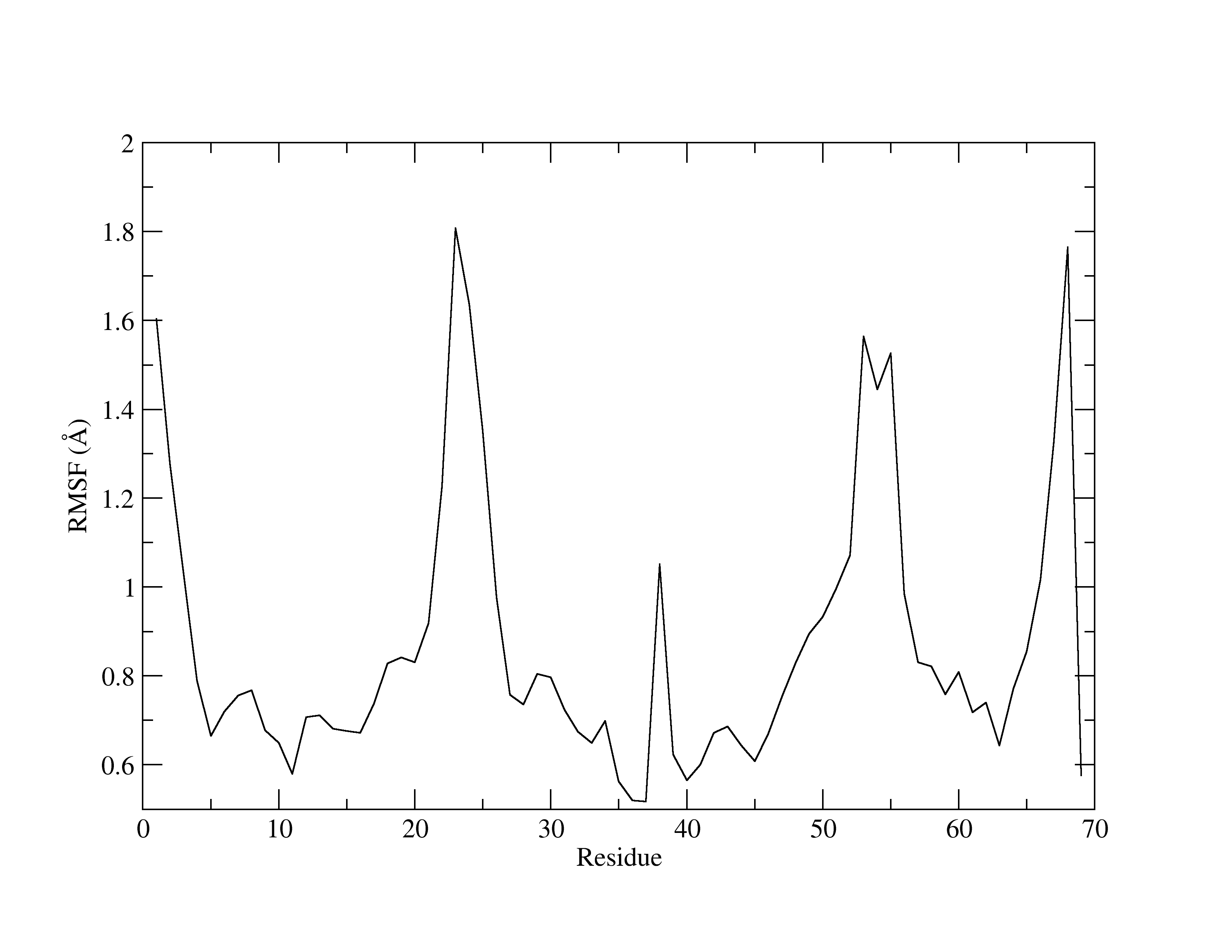
Figure 32. RMSF of MTR1.
The x-axis is the residue number, while the y-axis shows the fluctuations (in Å). Higher peaks indicate greater fluctuations. RMSFs are useful for identifying residues that have a greater contribution to the overall dynamics of the structure. Usually, it is common for residues involved in crystal contacts to exhibit higher peaks, reflecting increased mobility.
Lastly, you will learn to obtain the average structure of a trajectory using CPPTRAJ with the command average.
Find the input file ctraj.average_MTR1.in and VIM it.
parm MTR1_prot-run.parm7
trajin centered.MTR1_prot-run.nc
average avg.pdb :1-69&!@H= pdb
run
quit
After the average command, you must provide a filename in which to save the data. Here, the name is avg.pdb, followed by the atom selection mask and the file type specification pdb.
Run the input file as usual:
[user@expanse] cpptraj -i ctraj.average_MTR1.in
Now, you have an average structure of MTR1 in a PDB format that can be opened up in VMD for viewing.
In this tutorial, you have gained foundational skills in using VMD and CPPTRAJ for visualizing and analyzing trajectories from MD simulations. On the completion of this tutorial, you are ready to proceed to the more advanced AMBER tutorials ahead. Both VMD and CPPTRAJ can be very valuable tools for your studies, so continue to practice and even venture into the more complex analyses that these powerful programs offer.