Hands-On Session 10: Perform AFE simulations for Absolute Binding, using free energy workflows
Learning objectives
Learn how to build simulation boxes for absolute binding free energy calculations (ABFE) using AmberTools
Understand the options for solvating simulation boxes and adding ions
Learn how to make consistent solvent boxes for both the bound and unbound states of the ligand.
Learn how to equilibrate the endstates in preparation for running absolute binding free energy calculations (ABFE).
Learn how to determine Boresch restraints from end-state trajectories.
Learn how to set up and generate lambda windows for absolute binding free energy (ABFE) calculations starting from an equilbrated end-state.
Learn how to run ABFE calculations using Amber
Learn how to analyze absolute binding free energy calculations using FE-Toolkit and Edgembar.
Understand how to interpret the results of ABFE calculations for the Tyk2 system.
Learn to use AmberFlow to set up and run absolute binding free energy calculations (ABFE) for a protein-ligand system.
Relevant literature
Outline
In this tutorial, we will be using the python package LigandParam, developed by the York Group, to parameterize a ligand for use in molecular dynamics simulations. For this tutorial, we will be parameterizing the ligand…
Tutorial
flowchart LR
Download["Download tutorial files<br>Inputs - build_files.tar.gz<br>Outputs - extracted build_files"]
subgraph Aqueous_Leg ["Aqueous Phase"]
direction TB
AL1["Load ligand files and leaprc<br>Inputs - binder_XXX.lib binder_XXX.frcmod binder_XXX.pdb"]
AL2["Build prebox with tleap<br>Commands - setbox centers<br>addions2 Na+ 0 Cl- 0<br>solvateoct TIP4PBOX 16.0<br>Outputs - prebox_binder_XXX.parm7 .rst7"]
AL3["Count waters and compute ions<br>Subprogram - count_waters.py<br>Calc - N_ions = N_water * 0.150 / 55<br>Outputs - N ions per type"]
AL4["Add ions and save final binder<br>Command - addionsrand mol Na+ 7 Cl- 7 6.0 (example)<br>Outputs - binder_XXX.parm7 .rst7"]
end
subgraph Complex_Leg ["Bound Complex"]
direction TB
CL1["Load ligand and protein files and leaprc<br>Inputs - binder_XXX.lib binder_XXX.frcmod binder_XXX.pdb target.pdb"]
CL2["Combine and build prebox with tleap<br>Commands - combine mol prot<br>setbox centers<br>solvateoct TIP4PBOX 14.0<br>Outputs - prebox_target_XXX.parm7 .rst7"]
CL3["Count waters and compute ions for complex<br>Subprogram - count_waters.py<br>Example - 11023 waters -> 30 Na+ 30 Cl-<br>Outputs - N ions per type"]
CL4["Add ions to complex and save final system<br>Command - addionsrand complex Na+ 30 Cl- 30 6.0 (example)<br>Outputs - target_XXX.parm7 .rst7"]
end
%% Parallel legs from the common Download node
Download --> AL1
Download --> CL1
AL1 --> AL2
CL1 --> CL2
AL2 --> AL3
CL2 --> CL3
AL3 --> AL4
CL3 --> CL4
%% Join the two legs into the optional HMR step
AL4 --> HMR["Optional Hydrogen Mass Repartitioning<br>Tool - ParmEd<br>Commands - parm file<br>hmassrepartition<br>outparm *_hmr.parm7<br>Outputs - *_hmr.parm7"]
CL4 --> HMR
HMR --> END["Ready for minimization equilibration and ABFE workflows<br>Notes - repeat for each ligand; keep solvent box consistency if desired"]
%% End of flowchart
%% End of flowchart
Building Simulation Boxes for ABFE Calculations
To get started, you will need to download the tutorial files. The files include pdbs for the protein, and pdbs with docked coordinates for the ligand. The files in this download also include pre-generated forcefield parameters for each of the four ligands, which you will use in this tutorial.
Once you have downloaded the files, extract them into a directory where you will be working on this tutorial.
tar -xvzf build_files.tar.gz
cd build_files
Building the Ligand in the Aqueous Phase
In this section, you will build the ligand in a solvent box. You will be using tleap to build the system.
For this tutorial, we will be starting with the ligand ejm_31 and doing both builds for the bound and unbound states with this ligand. We will then be using the same procudure for the other three ligands. For each code block below, we have provided tabs for the other ligands so that you can easily switch between them.
Warning
In this tutorial, we will not be making a significant effort to enforce best practices for preparing the solvent box. In a production settign, you likely will desire to enforce that the solvent box has the same number of water molecules for all ligands, and depending on the identity of the ligand you may want to use a different box size or shape.
To build the ligand in a solvent box, you will need to create a tleap input file.
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_ejm_31.lib
loadamberparams ../binder_ejm_31.frcmod
mol = loadpdb ../binder_ejm_31.pdb
setbox mol centers
addions2 mol Na+ 0
addions2 mol Cl- 0
solvateoct mol TIP4PBOX 16.0
saveamberparm mol prebox_binder_ejm_31.parm7 prebox_binder_ejm_31.rst7
quit
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_ejm_42.lib
loadamberparams ../binder_ejm_42.frcmod
mol = loadpdb ../binder_ejm_42.pdb
setbox mol centers
addions2 mol Na+ 0
addions2 mol Cl- 0
solvateoct mol TIP4PBOX 16.0
saveamberparm mol prebox_binder_ejm_42.parm7 prebox_binder_ejm_42.rst7
quit
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew.. tab-set::
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_jmc_27.lib
loadamberparams ../binder_jmc_27.frcmod
mol = loadpdb ../binder_jmc_27.pdb
setbox mol centers
addions2 mol Na+ 0
addions2 mol Cl- 0
solvateoct mol TIP4PBOX 16.0
saveamberparm mol prebox_binder_jmc_27.parm7 prebox_binder_jmc_27.rst7
quit
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_jmc_28.lib
loadamberparams ../binder_jmc_28.frcmod
mol = loadpdb ../binder_jmc_28.pdb
setbox mol centers
addions2 mol Na+ 0
addions2 mol Cl- 0
solvateoct mol TIP4PBOX 16.0
saveamberparm mol prebox_binder_jmc_28.parm7 prebox_binder_jmc_28.rst7
quit
To run any of these scripts, save the desired script (e.g., tleap_ejm_31.in) and then run the following command:
tleap -f tleap_ejm_31.in
Calculating the Number of Ions to Add
If everything runs correctly, you should see two new files in your directory. We’ve called these prebox_binder_XXX.parm7 and prebox_binder_XXX.rst7, where XXX is the ligand name. These files contain the topology and coordinates for the ligand in a solvent box, with any neutralizing ions added. Now, if we were happy without ions, we could move on to minimization and equilibration of this system. However, we want to add ions to the system to mimic a 150 mM NaCl concentration.
To do this, we will first need to calculate the number of ions to add. First, we must find the number of water molecules in the box. We can use a quick MDAnalysis script to do this.
Save the following script as count_waters.py:
import MDAnalysis as mda
import sys
u = mda.Universe(sys.argv[1], sys.argv[2], format="INPCRD")
waters = u.select_atoms("resname WAT")
print(f"Number of water molecules: {len(waters.residues)}")
Then run the script as follows:
python count_waters.py prebox_binder_ejm_31.parm7 prebox_binder_ejm_31.rst7
python count_waters.py prebox_binder_ejm_42.parm7 prebox_binder_ejm_42.rst7
python count_waters.py prebox_binder_jmc_27.parm7 prebox_binder_jmc_27.rst7
python count_waters.py prebox_binder_jmc_28.parm7 prebox_binder_jmc_28.rst7
Each of these will output a number of water molecules in the box. We will then use these to calculate the number of ions to add.
The way we do this calculation is to use the fact that water is 55 M, so we can use the following formula to calculate the number of ions to add:
When running for this tutorial, we found that we had 2,470 water molecules in the box for ejm_31, which gives us 7 ions of each type to add. For simplicity, we will be adding 7 ions of each type to each of the systems in this tutorial.
Now we can write a new tleap script to add the ions and save the final topology and coordinates.
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_ejm_31.lib
loadamberparams ../binder_ejm_31.frcmod
mol = loadpdb ../binder_ejm_31.pdb
setbox mol centers
addions2 mol Na+ 0
addions2 mol Cl- 0
solvateoct mol TIP4PBOX 16.0
addionsrand mol Na+ 7 Cl- 7 6.0
saveamberparm mol binder_ejm_31.parm7 binder_ejm_31.rst7
quit
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_ejm_42.lib
loadamberparams ../binder_ejm_42.frcmod
mol = loadpdb ../binder_ejm_42.pdb
setbox mol centers
addions2 mol Na+ 0
addions2 mol Cl- 0
solvateoct mol TIP4PBOX 16.0
addionsrand mol Na+ 7 Cl- 7 6.0
saveamberparm mol binder_ejm_42.parm7 binder_ejm_42.rst7
quit
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_jmc_27.lib
loadamberparams ../binder_jmc_27.frcmod
mol = loadpdb ../binder_jmc_27.pdb
setbox mol centers
addions2 mol Na+ 0
addions2 mol Cl- 0
solvateoct mol TIP4PBOX 16.0
addionsrand mol Na+ 7 Cl- 7 6.0
saveamberparm mol binder_jmc_27.parm7 binder_jmc_27.rst7
quit
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_jmc_28.lib
loadamberparams ../binder_jmc_28.frcmod
mol = loadpdb ../binder_jmc_28.pdb
setbox mol centers
addions2 mol Na+ 0
addions2 mol Cl- 0
solvateoct mol TIP4PBOX 16.0
addionsrand mol Na+ 7 Cl- 7 6.0
saveamberparm mol binder_jmc_28.parm7 binder_jmc_28.rst7
quit
To run any of these scripts, save the desired script (e.g., tleap_ejm_31.in) and then run the following command:
tleap -f tleap_ejm_31.in
Again, these scripts can be run as follows:
tleap -f tleap_ejm_31_addions.in
tleap -f tleap_ejm_42_addions.in
tleap -f tleap_jmc_27_addions.in
tleap -f tleap_jmc_28_addions.in
There we go! We now have the ligand built in a solvent box with ions added. You should see two new files in your directory called binder_XXX.parm7 and binder_XXX.rst7, where XXX is the ligand name. These files contain the topology and coordinates for the ligand in a solvent box, with 150 mM NaCl added.
Building the Bound Protein-Ligand Complex
Now, we will build the bound protein-ligand complex. The procedure is very similar to what we did for the ligand in a solvent box, but now we will be loading in the protein as well.
We will again be using tleap to build the system; however, this time we will be loading in the protein pdb as well as the ligand pdb.
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_ejm_31.lib
loadamberparams ../binder_ejm_31.frcmod
mol = loadpdb ../binder_ejm_31.pdb
prot = loadpdb ../target.pdb
complex = combine {mol prot}
setbox complex centers
addions2 complex Na+ 0
addions2 complex Cl- 0
solvateoct complex TIP4PBOX 14.0
saveamberparm complex prebox_target_ejm_31.parm7 prebox_target_ejm_31.rst7
quit
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_ejm_42.lib
loadamberparams ../binder_ejm_42.frcmod
mol = loadpdb ../binder_ejm_42.pdb
prot = loadpdb ../target.pdb
complex = combine {mol prot}
setbox complex centers
addions2 complex Na+ 0
addions2 complex Cl- 0
solvateoct complex TIP4PBOX 14.0
saveamberparm complex prebox_target_ejm_42.parm7 prebox_target_ejm_42.rst7
quit
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_jmc_27.lib
loadamberparams ../binder_jmc_27.frcmod
mol = loadpdb ../binder_jmc_27.pdb
prot = loadpdb ../target.pdb
complex = combine {mol prot}
setbox complex centers
addions2 complex Na+ 0
addions2 complex Cl- 0
solvateoct complex TIP4PBOX 14.0
saveamberparm complex prebox_target_jmc_27.parm7 prebox_target_jmc_27.rst7
quit
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_jmc_28.lib
loadamberparams ../binder_jmc_28.frcmod
mol = loadpdb ../binder_jmc_28.pdb
prot = loadpdb ../target.pdb
complex = combine {mol prot}
setbox complex centers
addions2 complex Na+ 0
addions2 complex Cl- 0
solvateoct complex TIP4PBOX 14.0
saveamberparm complex prebox_target_jmc_28.parm7 prebox_target_jmc_28.rst7
quit
To run any of these scripts, save the desired script (e.g., tleap_target_ejm_31.in) and then run the following command:
tleap -f tleap_target_ejm_31.in
Again, if everything runs correctly, you should see two new files in your directory. We’ve called these prebox_target_XXX.parm7 and prebox_target_XXX.rst7, where XXX is the ligand name. These files contain the topology and coordinates for the protein-ligand complex in a solvent box, with any neutralizing ions added. Now, if we were happy without ions, we could move on to minimization and equilibration of this system. However, we want to add ions to the system to mimic a 150 mM NaCl concentration.
Just like before, we will use the count_waters.py script to find the number of water molecules in the box.
Run the script as follows:
python count_waters.py prebox_target_ejm_31.parm7 prebox_target_ejm_31.rst7
python count_waters.py prebox_target_ejm_42.parm7 prebox_target_ejm_42.rst7
python count_waters.py prebox_target_jmc_27.parm7 prebox_target_jmc_27.rst7
python count_waters.py prebox_target_jmc_28.parm7 prebox_target_jmc_28.rst7
In this case, we found that we had 11,023 water molecules in the box for ejm_31, which gives us 30 ions of each type to add. For simplicity, we will be adding 30 ions of each type to each of the systems in this tutorial.
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_ejm_31.lib
loadamberparams ../binder_ejm_31.frcmod
mol = loadpdb ../binder_ejm_31.pdb
prot = loadpdb ../target.pdb
complex = combine {mol prot}
setbox complex centers
addions2 complex Na+ 0
addions2 complex Cl- 0
solvateoct complex TIP4PBOX 14.0
addionsrand complex Na+ 30 Cl- 30 6.0
saveamberparm complex target_ejm_31.parm7 target_ejm_31.rst7
quit
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_ejm_42.lib
loadamberparams ../binder_ejm_42.frcmod
mol = loadpdb ../binder_ejm_42.pdb
prot = loadpdb ../target.pdb
complex = combine {mol prot}
setbox complex centers
addions2 complex Na+ 0
addions2 complex Cl- 0
solvateoct complex TIP4PBOX 14.0
addionsrand complex Na+ 30 Cl- 30 6.0
saveamberparm complex target_ejm_42.parm7 target_ejm_42.rst7
quit
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_jmc_27.lib
loadamberparams ../binder_jmc_27.frcmod
mol = loadpdb ../binder_jmc_27.pdb
prot = loadpdb ../target.pdb
complex = combine {mol prot}
setbox complex centers
addions2 complex Na+ 0
addions2 complex Cl- 0
solvateoct complex TIP4PBOX 14.0
addionsrand complex Na+ 30 Cl- 30 6.0
saveamberparm complex target_jmc_27.parm7 target_jmc_27.rst7
quit
source leaprc.protein.ff19SB
source leaprc.phosaa19SB
loadamberparams frcmod.ff19SB
source leaprc.gaff2
source leaprc.water.tip4pew
loadamberparams frcmod.tip4pew
loadamberparams frcmod.ionsjc_tip4pew
loadoff ../binder_jmc_28.lib
loadamberparams ../binder_jmc_28.frcmod
mol = loadpdb ../binder_jmc_28.pdb
prot = loadpdb ../target.pdb
complex = combine {mol prot}
setbox complex centers
addions2 complex Na+ 0
addions2 complex Cl- 0
solvateoct complex TIP4PBOX 14.0
addionsrand complex Na+ 30 Cl- 30 6.0
saveamberparm complex target_jmc_28.parm7 target_jmc_28.rst7
quit
As with above, to run any of these scripts, save the desired script (e.g., tleap_target_ejm_31_addions.in) and then run the following command:
tleap -f tleap_target_ejm_31_addions.in
tleap -f tleap_target_ejm_42_addions.in
tleap -f tleap_target_jmc_27_addions.in
tleap -f tleap_target_jmc_28_addions.in
Thus, now we have topology files for the ligand in complex for all four ligands! These ligands can now be used for further simulations and analysis.
Optional: Applying Hydrogen Mass Reparationing (HMR)
Often, in order to speed up simulations, we will apply hydrogen mass repartitioning (HMR) to the system. This allows us to use a larger timestep (4 fs vs 2 fs) for production simulations.
To do this, we will be using the ParmEd package that comes with AmberTools. The following command will apply HMR to a given topology and coordinate file, and output new files with HMR applied.
We will need to make a parmed input file for each of the systems. Here is an example for ejm_31 in complex:
parm target_ejm_31.parm7
hmassrepartition
outparm target_ejm_31_hmr.parm7
parm target_ejm_42.parm7
hmassrepartition
outparm target_ejm_42_hmr.parm7
parm target_jmc_27.parm7
hmassrepartition
outparm target_jmc_27_hmr.parm7
parm target_jmc_28.parm7
hmassrepartition
outparm target_jmc_28_hmr.parm7
The same can be done for the binder:
parm binder_ejm_31.parm7
hmassrepartition
outparm binder_ejm_31_hmr.parm7
parm binder_ejm_42.parm7
hmassrepartition
outparm binder_ejm_42_hmr.parm7
parm binder_jmc_27.parm7
hmassrepartition
outparm binder_jmc_27_hmr.parm7
parm binder_jmc_28.parm7
hmassrepartition
outparm binder_jmc_28_hmr.parm7
These files can be run using the following command:
parmed -i parmed_ejm_31.in
For simplicity, a script to do this for all four ligands is provided below:
for ligand in ejm_31 ejm_42 jmc_27 jmc_28; do
echo "parm target_${ligand}.parm7" > parmed_${ligand}_target.in
echo "hmassrepartition" >> parmed_${ligand}_target.in
echo "outparm target_${ligand}_hmr.parm7" >> parmed_${ligand}_target.in
echo "parm binder_${ligand}.parm7" > parmed_${ligand}_binder.in
echo "hmassrepartition" >> parmed_${ligand}_binder.in
echo "outparm binder_${ligand}_hmr.parm7" >> parmed_${ligand}_binder.in
parmed -i parmed_${ligand}_target.in
parmed -i parmed_${ligand}_binder.in
done
flowchart LR
%% Aqueous endstate boxed path
subgraph AQ[Aqueous Endstate Equilibration]
direction LR
A0[Aqueous Start<br>Inputs: binder parm7 and rst7<br>Outputs: Initial aqueous rst7] --> A1
A1[Minimization<br>Inputs: binder rst7<br>Subprograms: pmemd minimization.mdin<br>Outputs: binder minimization rst7] --> A2
A2[Heating<br>Inputs: minimized aqueous rst7<br>Subprograms: pmemd heat.mdin<br>Outputs: heated aqueous rst7] --> A3
A3[Density 1<br>Inputs: heated aqueous rst7<br>Subprograms: pmemd density1.mdin<br>Outputs: density1 aqueous rst7] --> A4
A4[Density 2<br>Inputs: density1 aqueous rst7<br>Subprograms: pmemd density2.mdin<br>Outputs: density2 aqueous rst7] --> A5
A5[Lower Restraints 1<br>Inputs: density2 aqueous rst7<br>Subprograms: pmemd lower_restraints1.mdin<br>Outputs: restrained aqueous rst7] --> A6
A6[Lower Restraints 2<br>Inputs: restrained aqueous rst7<br>Subprograms: pmemd lower_restraints2.mdin<br>Outputs: restrained aqueous rst7] --> A7
A7[Lower Restraints 3<br>Inputs: restrained aqueous rst7<br>Subprograms: pmemd lower_restraints3.mdin<br>Outputs: restrained aqueous rst7] --> A8
A8[Lower Restraints 4<br>Inputs: restrained aqueous rst7<br>Subprograms: pmemd lower_restraints4.mdin<br>Outputs: restrained aqueous rst7] --> A9
A9[Lower Restraints 5<br>Inputs: restrained aqueous rst7<br>Subprograms: pmemd lower_restraints5.mdin<br>Outputs: lightly restrained aqueous rst7] --> A10
A10[Backbone Binder Restraints<br>Inputs: lightly restrained aqueous rst7<br>Subprograms: pmemd backbone.mdin<br>Outputs: backbone restrained aqueous rst7] --> A11
A11[Unrestrained MD<br>Inputs: backbone restrained aqueous rst7<br>Subprograms: pmemd free.mdin<br>Outputs: equilibrated aqueous rst7]
end
%% Complex endstate boxed path
subgraph CX[Complex Endstate Equilibration]
direction LR
B0[Complex Start<br>Inputs: target parm7 and rst7<br>Outputs: Initial complex rst7] --> B1
B1[Minimization<br>Inputs: complex rst7<br>Subprograms: pmemd minimization.mdin<br>Outputs: complex minimization rst7] --> B2
B2[Heating<br>Inputs: minimized complex rst7<br>Subprograms: pmemd heat.mdin with protein and ligand restraints<br>Outputs: heated complex rst7] --> B3
B3[Density 1<br>Inputs: heated complex rst7<br>Subprograms: pmemd density1.mdin<br>Outputs: density1 complex rst7] --> B4
B4[Density 2<br>Inputs: density1 complex rst7<br>Subprograms: pmemd density2.mdin<br>Outputs: density2 complex rst7] --> B5
B5[Lower Restraints 1<br>Inputs: density2 complex rst7<br>Subprograms: pmemd lower_restraints1.mdin<br>Outputs: restrained complex rst7] --> B6
B6[Lower Restraints 2<br>Inputs: restrained complex rst7<br>Subprograms: pmemd lower_restraints2.mdin<br>Outputs: restrained complex rst7] --> B7
B7[Lower Restraints 3<br>Inputs: restrained complex rst7<br>Subprograms: pmemd lower_restraints3.mdin<br>Outputs: restrained complex rst7] --> B8
B8[Lower Restraints 4<br>Inputs: restrained complex rst7<br>Subprograms: pmemd lower_restraints4.mdin<br>Outputs: restrained complex rst7] --> B9
B9[Lower Restraints 5<br>Inputs: restrained complex rst7<br>Subprograms: pmemd lower_restraints5.mdin<br>Outputs: lightly restrained complex rst7] --> B10
B10[Backbone Binder Restraints<br>Inputs: lightly restrained complex rst7<br>Subprograms: pmemd backbone.mdin<br>Outputs: backbone restrained complex rst7] --> B11
B11[Unrestrained MD<br>Inputs: backbone restrained complex rst7<br>Subprograms: pmemd free.mdin<br>Outputs: equilibrated complex rst7]
end
OUT['Final Equilibrated Systems']
A11 --> OUT
B11 --> OUT
Setting Up and Running End-State Equilibration
The starting point for this tutorial is the topology (parm7) and coordinate (rst7) files for the complex and aqueous endstates. The files for this tutorial are located:
Once you have downloaded the files, extract them into a directory where you will be working on this tutorial.
tar -xvzf equil_tyk2.tar.gz
cd equil_tyk2
mkdir working_dir
cd working_dir
The files you should see in the top level directory are parm7 and rst7 files for both the complex and aqueous end states. The parm7 files contain the hmr tag indicating that hydrogen mass repartitioning has been used to increase the timestep of the simulations to 4 fs.
Note
While this tutorial focuses on equilibrating for ABFE calculations, these are generally good practices for equilibrating any protein system using Amber.
Equilibrating the Aqueous Endstates
To equilibrate the aqueous endstates, we will be using a number of steps to gradually relax the system, the MD input files are included below, and again can be found in the tutorial files you downloaded above.
&cntrl
imin = 1, maxcyc = 1000,
ncyc = 100, ntx = 1, ntmin = 1,
ntwe = 0, ntwr = 0, ntpr = 1000,
ntc = 2, ntf = 2, ntb = 1, ntp = 0,
cut = 10.0, ntr = 1, restraintmask = ':1&!@H=', restraint_wt = 50,
ioutfm=1, ntxo=1,
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.001
irest = 0
ntx = 1
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 250
ntwe = 0
ntwr = 250
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 1.0
ntb = 1
ntp = 0
barostat = 2
ntr=1
restraint_wt= 50
restraintmask=':1&!@H='
nmropt=1
/
&wt TYPE="TEMP0"
istep1=0
istep2=1000000
value1=298
value2=298
/
&wt TYPE="END"
/
&cntrl
imin = 0
nstlim = 20000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 2.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 50
restraintmask=':1&!@H='
/
&cntrl
imin = 0
nstlim = 20000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 2.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 50
restraintmask=':1&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 4.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 25
restraintmask=':1&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 4.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 10
restraintmask=':1&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 4.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 5
restraintmask=':1&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 4.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 2
restraintmask=':1&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 4.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 1
restraintmask=':1&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.002
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 250
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 5.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 0.5
restraintmask='@CA,N,C | :1&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.004,
irest = 1
ntx = 5
ig = -1,
ntc = 2
ntf = 2
tol = 1e-05,
nscm = 0
ioutfm=1
ntxo=1,
ntwx = 250
ntwe = 0
ntwr = 500
ntpr = 1000,
cut = 10.0
iwrap = 0,
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 5.0
ntb = 2
ntp = 1,
barostat = 2,
/
There are a total of 11 steps to equilibrate the aqueous endstate; however, many of these steps can be grouped together into general goals.
These goals are:
Minimize the system to remove bad contacts (step 1)
Heat the system to the target temperature while restraining the solute (step 2)
Equilibrate the density of the system while restraining the solute (steps 3 and 4)
Gradually relax the solute restraints (steps 5-10)
Run a short unrestrained simulation to relax the system (step 11)
Below, we have included the commands to run each of these steps using amber.
pmemd.cuda -O -i ../binder_mdins/minimization.mdin -c ../binder_ejm_31.rst7 -p ../binder_ejm_31_hmr.parm7 -x binder_ejm_31_minimization.nc -r binder_ejm_31_minimization.rst7 -o binder_ejm_31_minimization.mdout -ref ../binder_ejm_31.rst7
pmemd.cuda -O -i ../binder_mdins/heat.mdin -c binder_ejm_31_minimization.rst7 -p ../binder_ejm_31_hmr.parm7 -x binder_ejm_31_heat.nc -r binder_ejm_31_heat.rst7 -o binder_ejm_31_heat.mdout -ref ../binder_ejm_31.rst7
pmemd.cuda -O -i ../binder_mdins/density1.mdin -c binder_ejm_31_heat.rst7 -p ../binder_ejm_31_hmr.parm7 -x binder_ejm_31_density1.nc -r binder_ejm_31_density1.rst7 -o binder_ejm_31_density1.mdout -ref ../binder_ejm_31.rst7
pmemd.cuda -O -i ../binder_mdins/density2.mdin -c binder_ejm_31_density1.rst7 -p ../binder_ejm_31_hmr.parm7 -x binder_ejm_31_density2.nc -r binder_ejm_31_density2.rst7 -o binder_ejm_31_density2.mdout -ref ../binder_ejm_31.rst7
pmemd.cuda -O -i ../binder_mdins/lower_restraints1.mdin -c binder_ejm_31_density2.rst7 -p ../binder_ejm_31_hmr.parm7 -x binder_ejm_31_lower_restraints1.nc -r binder_ejm_31_lower_restraints1.rst7 -o binder_ejm_31_lower_restraints1.mdout -ref ../binder_ejm_31.rst7
pmemd.cuda -O -i ../binder_mdins/lower_restraints2.mdin -c binder_ejm_31_lower_restraints1.rst7 -p ../binder_ejm_31_hmr.parm7 -x binder_ejm_31_lower_restraints2.nc -r binder_ejm_31_lower_restraints2.rst7 -o binder_ejm_31_lower_restraints2.mdout -ref ../binder_ejm_31.rst7
pmemd.cuda -O -i ../binder_mdins/lower_restraints3.mdin -c binder_ejm_31_lower_restraints2.rst7 -p ../binder_ejm_31_hmr.parm7 -x binder_ejm_31_lower_restraints3.nc -r binder_ejm_31_lower_restraints3.rst7 -o binder_ejm_31_lower_restraints3.mdout -ref ../binder_ejm_31.rst7
pmemd.cuda -O -i ../binder_mdins/lower_restraints4.mdin -c binder_ejm_31_lower_restraints3.rst7 -p ../binder_ejm_31_hmr.parm7 -x binder_ejm_31_lower_restraints4.nc -r binder_ejm_31_lower_restraints4.rst7 -o binder_ejm_31_lower_restraints4.mdout -ref ../binder_ejm_31.rst7
pmemd.cuda -O -i ../binder_mdins/lower_restraints5.mdin -c binder_ejm_31_lower_restraints4.rst7 -p ../binder_ejm_31_hmr.parm7 -x binder_ejm_31_lower_restraints5.nc -r binder_ejm_31_lower_restraints5.rst7 -o binder_ejm_31_lower_restraints5.mdout -ref ../binder_ejm_31.rst7
pmemd.cuda -O -i ../binder_mdins/backbone.mdin -c binder_ejm_31_lower_restraints5.rst7 -p ../binder_ejm_31_hmr.parm7 -x binder_ejm_31_backbone.nc -r binder_ejm_31_backbone.rst7 -o binder_ejm_31_backbone.mdout -ref ../binder_ejm_31.rst7
pmemd.cuda -O -i ../binder_mdins/free.mdin -c binder_ejm_31_backbone.rst7 -p ../binder_ejm_31_hmr.parm7 -x binder_ejm_31_unrestrained.nc -r binder_ejm_31_unrestrained.rst7 -o binder_ejm_31_unrestrained.mdout -ref ../binder_ejm_31.rst7
Note
On Running Amber commands
The above commands use the pmemd.cuda executable to run the simulations on a GPU. If you are running on a CPU, you can replace pmemd.cuda with pmemd.MPI or sander as appropriate.
Every command uses a number of common flags, which we will break down below for the first pmemd.cuda command.
pmemd.cuda -O -i ../binder_mdins/minimization.mdin -c ../binder_ejm_31.rst7 -p ../binder_ejm_31_hmr.parm7 -x binder_ejm_31_minimization.nc -r binder_ejm_31_minimization.rst7 -o binder_ejm_31_minimization.mdout -ref ../binder_ejm_31.rst7
The flags used here are:
-O: Overwrite output files if they already exist
-i: Input mdin file
-c: Input coordinate file (rst7)
-p: Input topology file (parm7)
-x: Output trajectory file (nc)
-r: Output restart file (rst7)
-o: Output mdout file (text)
-ref: Reference coordinate file for restraints
In the mdin files, the restraints are defined using the reference coordinate file provided with the -ref flag. We are currently using Root Mean Square (RMS) restrains on the ligand heavy atoms to keep them close to this reference structure during equilibration.
Equilibrating the Complex Endstates
To equilibrate the complex endstates, we will follow essentially the same steps as we did for the aqueous endstate, with the same general goals. The primary difference between these mdins and the previous is the protein restraint mask 1-289, which restrains the protein residues in addition to the ligand. The mdin files are included below, and again can be found in the tutorial files you downloaded above.
Warning
Note that the mdin restrains here have been tailored to the TYK2 system, and should be adjusted if you are working with a different system. The key change to make is to adjust the protein residue range in the restraint mask (e.g. 1-289) to match the protein you are working with, plus 1 to account for the ligand being the first residue.
&cntrl
imin = 1, maxcyc = 1000,
ncyc = 100, ntx = 1, ntmin = 1,
ntwe = 0, ntwr = 0, ntpr = 1000,
ntc = 2, ntf = 2, ntb = 1, ntp = 0,
cut = 10.0, ntr = 1, restraintmask = ':1-289&!@H=', restraint_wt = 50,
ioutfm=1, ntxo=1,
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.001
irest = 0
ntx = 1
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 250
ntwe = 0
ntwr = 250
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 1.0
ntb = 1
ntp = 0
barostat = 2
ntr=1
restraint_wt= 50
restraintmask=':1-289&!@H='
nmropt=1
/
&wt TYPE="TEMP0"
istep1=0
istep2=1000000
value1=298
value2=298
/
&wt TYPE="END"
/
&cntrl
imin = 0
nstlim = 20000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 2.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 50
restraintmask=':1-289&!@H='
/
&cntrl
imin = 0
nstlim = 20000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 2.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 50
restraintmask=':1-289&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 4.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 25
restraintmask=':1-289&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 4.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 10
restraintmask=':1-289&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 4.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 5
restraintmask=':1-289&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 4.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 2
restraintmask=':1-289&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.001
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 0
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 4.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 1
restraintmask=':1-289&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.002
irest = 1
ntx = 5
ig = -1
ntc = 2
ntf = 2
tol = 1e-05
nscm = 0
ioutfm=1
ntxo=1
ntwx = 250
ntwe = 0
ntwr = 500
ntpr = 1000
cut = 10.0
iwrap = 0
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 5.0
ntb = 2
ntp = 1
barostat = 2
ntr=1
restraint_wt= 0.5
restraintmask='@CA,N,C | :1&!@H='
/
&cntrl
imin = 0
nstlim = 1000000
dt = 0.004,
irest = 1
ntx = 5
ig = -1,
ntc = 2
ntf = 2
tol = 1e-05,
nscm = 0
ioutfm=1
ntxo=1,
ntwx = 250
ntwe = 0
ntwr = 500
ntpr = 1000,
cut = 10.0
iwrap = 0,
ntt = 3
temp0 = 298
tempi = 298 ! tempi is the initial temperature, temp0 is the target
gamma_ln = 5.0
ntb = 2
ntp = 1,
barostat = 2,
/
The commands to run each of these steps using amber are included below.
pmemd.cuda -O -i ../complex_mdins/minimization.mdin -c ../target_ejm_31.rst7 -p ../target_ejm_31_hmr.parm7 -x complex_ejm_31_minimization.nc -r complex_ejm_31_minimization.rst7 -o complex_ejm_31_minimization.mdout -ref ../target_ejm_31.rst7
pmemd.cuda -O -i ../complex_mdins/heat.mdin -c complex_ejm_31_minimization.rst7 -p ../target_ejm_31_hmr.parm7 -x complex_ejm_31_heat.nc -r complex_ejm_31_heat.rst7 -o complex_ejm_31_heat.mdout -ref ../target_ejm_31.rst7
pmemd.cuda -O -i ../complex_mdins/density1.mdin -c complex_ejm_31_heat.rst7 -p ../target_ejm_31_hmr.parm7 -x complex_ejm_31_density1.nc -r complex_ejm_31_density1.rst7 -o complex_ejm_31_density1.mdout -ref ../target_ejm_31.rst7
pmemd.cuda -O -i ../complex_mdins/density2.mdin -c complex_ejm_31_density1.rst7 -p ../target_ejm_31_hmr.parm7 -x complex_ejm_31_density2.nc -r complex_ejm_31_density2.rst7 -o complex_ejm_31_density2.mdout -ref ../target_ejm_31.rst7
pmemd.cuda -O -i ../complex_mdins/lower_restraints1.mdin -c complex_ejm_31_density2.rst7 -p ../target_ejm_31_hmr.parm7 -x complex_ejm_31_lower_restraints1.nc -r complex_ejm_31_lower_restraints1.rst7 -o complex_ejm_31_lower_restraints1.mdout -ref ../target_ejm_31.rst7
pmemd.cuda -O -i ../complex_mdins/lower_restraints2.mdin -c complex_ejm_31_lower_restraints1.rst7 -p ../target_ejm_31_hmr.parm7 -x complex_ejm_31_lower_restraints2.nc -r complex_ejm_31_lower_restraints2.rst7 -o complex_ejm_31_lower_restraints2.mdout -ref ../target_ejm_31.rst7
pmemd.cuda -O -i ../complex_mdins/lower_restraints3.mdin -c complex_ejm_31_lower_restraints2.rst7 -p ../target_ejm_31_hmr.parm7 -x complex_ejm_31_lower_restraints3.nc -r complex_ejm_31_lower_restraints3.rst7 -o complex_ejm_31_lower_restraints3.mdout -ref ../target_ejm_31.rst7
pmemd.cuda -O -i ../complex_mdins/lower_restraints4.mdin -c complex_ejm_31_lower_restraints3.rst7 -p ../target_ejm_31_hmr.parm7 -x complex_ejm_31_lower_restraints4.nc -r complex_ejm_31_lower_restraints4.rst7 -o complex_ejm_31_lower_restraints4.mdout -ref ../target_ejm_31.rst7
pmemd.cuda -O -i ../complex_mdins/lower_restraints5.mdin -c complex_ejm_31_lower_restraints4.rst7 -p ../target_ejm_31_hmr.parm7 -x complex_ejm_31_lower_restraints5.nc -r complex_ejm_31_lower_restraints5.rst7 -o complex_ejm_31_lower_restraints5.mdout -ref ../target_ejm_31.rst7
pmemd.cuda -O -i ../complex_mdins/backbone.mdin -c complex_ejm_31_lower_restraints5.rst7 -p ../target_ejm_31_hmr.parm7 -x complex_ejm_31_backbone.nc -r complex_ejm_31_backbone.rst7 -o complex_ejm_31_backbone.mdout -ref ../target_ejm_31.rst7
pmemd.cuda -O -i ../complex_mdins/free.mdin -c complex_ejm_31_backbone.rst7 -p ../target_ejm_31_hmr.parm7 -x complex_ejm_31_unrestrained.nc -r complex_ejm_31_unrestrained.rst7 -o complex_ejm_31_unrestrained.mdout -ref ../target_ejm_31.rst7
Again, run these, and you will be left with equilibrated endstate files for both the complex and aqueous endstates, which can be used to start the production ABFE calculations. You can repeat these steps for each of the remaining three ligands; the mdin files are included in the tutorial files you downloaded above.
Determining Boresch Restraints
For this tutorial, we will be starting from the equilibrated end-state trajectory of a Tyk2-ligand complex in its bound state. These files were generated in the tutorial:ref:Equilibrating the End States of Tyk2 for ABFE Calculations <build_binder>; however, for convenience we have provided the necessary files below to get you started.
Attention
Update the download link to point to the finalized tutorial files once they are uploaded.
Theory of Boresch Restraints
Boresch restraints are a type of positional restraint used in molecular dynamics simulations to maintain the relative position and orientation of a ligand within a binding site. They are particularly useful in absolute binding free energy calculations, where it is essential to keep the ligand in place while decoupling its interactions with the environment.
The Boresch restraint method involves defining a set of six restraints based on three atoms from the ligand and three atoms from the protein. These restraints include:
One distance Restraint
Two Angle Restraints
Three Dihedral Restraints
These restraints work together to fix the ligand’s position and orientation relative to the protein, allowing for accurate free energy calculations.
Note
This theory section should be expanded with a more detailed explanation of the mathematical formulation of Boresch restraints, including equations and diagrams to illustrate the concepts.
Note
Much of the below theoretical discussion comes from Boresch et al., J. Phys. Chem. B, 2003, 107 (35), pp 9535–9551. DOI: 10.1021/jp0217839.
The goal of Boresch restraints is to define a known set of lambda-dependent restraints that can be applied to the ligand in the bound state to keep it in place while decoupling its interactions with the environment, while also allowing for an analytical correction to the free energy to account for the restraints.
Roughly, the situation that needs to be corrected for is:
Where here, (P cdot L)_{H_2O} is the protein-ligand complex in water with restraints applied, (P)_{H_2O} is the protein in water, and (L)_{H_2O} is the ligand in water. $Delta A_r$ is the free energy change associated with removing the restraints from the complex in water.
Because the ligand is in the dummy state when decoupled, the Ligand does not have interactions with the protein or solvent and thus the restraints are the only interactions with external sources it feels. The potential energy function for the system is:
Here, there are terms coming from the protein (\(U_P\)), the ligand (\(U_L\)), and the restraints (\(U_r\)).
The contribution to the free energy that comes from the restraints can be written as:
Where \(Z_{P \cdot L}\) is the partition function of the restrained complex, \(Z_P\) is the partition function of the protein, and \(Z_L\) is the partition function of the ligand.
This calculation is feasible analytically when the restraints are chosen such that
or in other words, choosing the restraints such that the partition function can be factored into independent contributions from the protein, ligand, and restraints.
After a few assumptions (Rigid Rotor) the final expression for the partition function is given by:
Finally, the free energy contribution from the restraints can be expressed as:
Where \(r_{aA,0}\) and \(\theta_{A,0}\) are the equilibrium distance and angle values for the restraints, and \(K_r\), \(K_{\theta^A}\), \(K_{\theta^B}\), \(K_{\phi^A}\), \(K_{\phi^B}\), and \(K_{\phi^C}\) are the force constants for the respective restraints.
How to Determine Boresch Restraints
To determine the Boresch restraints for your system, you will need to analyze an equilibrated trajectory for the protein-ligand complex.
Note
If you have been following along with the Tyk2 ABFE Tutorials, it is very likely that you have already generated a trajectory. It very likely that your trajectory will be different from the one included here, but the steps to generate the boresch restraints will be the same. Don’t be surprised if your final results differ from those presented here!
For this tutorial, we will be using the following python code, which you should save as generate_boresch_restraints.py:
#!/usr/bin/env python3
"""Generate Boresch restraints from equilibration trajectory.
This script scans ligand heavy atoms and nearby protein heavy atoms to
identify stable anchor pairs, computes Boresch degrees of freedom over
the trajectory, and prints/writes restraint parameters.
Behavior is preserved from the original script but reorganized for
readability and reuse.
"""
import argparse
import logging
from pathlib import Path
import MDAnalysis as mda
from MDAnalysis.analysis.distances import dist
from MDAnalysis.lib.distances import calc_dihedrals
import numpy as np
from numpy.linalg import norm
import matplotlib.pyplot as plt
LOG = logging.getLogger(__name__)
def parse_args():
"""Parse command-line arguments.
Returns
-------
argparse.Namespace
Object with attributes:
- topology: str, path to topology file (default './unisc.parm7')
- trajectory: str, path to trajectory file (default 'equil1/real_eq.nc')
- ligand_sel: str, MDAnalysis selection string for ligand heavy atoms
- cutoff: float, distance in angstroms to search for nearby protein atoms
- num_pairs: int, number of lowest-standard-deviation pairs to show/select
- max_pairs_traj: int, maximum number of pairs to analyze over the trajectory
- out_rest: str, output filename to write AMBER restraint input
- fig_prefix: str, prefix for any figures written
Notes
-----
This function only constructs and returns the parsed arguments object.
Documentation in tutorials should show running the script like:
$ python generate_boresch_restraints.py --topology my.parm7 --trajectory my.nc
"""
p = argparse.ArgumentParser(
description="Generate Boresch restraints from equilibration trajectory"
)
p.add_argument("--topology", type=str, default="./unisc.parm7", help="Topology file")
p.add_argument("--trajectory", type=str, default="equil1/real_eq.nc", help="Equilibration trajectory file")
p.add_argument("--ligand-sel", type=str, default="resid 1 and not name H*", help="Selection for ligand heavy atoms")
p.add_argument("--cutoff", type=float, default=10.0, help="Cutoff (angstrom) to search nearby protein atoms")
p.add_argument("--num-pairs", type=int, default=5, help="Number of low-SD pairs to show/select")
p.add_argument("--max-pairs-traj", type=int, default=200, help="Max pairs to analyze over trajectory")
p.add_argument("--out-rest", type=str, default="rest.in", help="Output restraint file")
p.add_argument("--fig-prefix", type=str, default="boresch", help="Prefix for saved figures")
return p.parse_args()
def find_anchor_pairs(u, ligand_sel="resid 1 and not name H*", cutoff=10.0):
"""Find candidate ligand-protein anchor atom pairs.
Parameters
----------
u : MDAnalysis.Universe
Loaded universe containing topology and trajectory.
ligand_sel : str
MDAnalysis selection string identifying ligand heavy atoms (no H).
cutoff : float
Distance cutoff (in Å) used to find protein heavy atoms near each
ligand heavy atom.
Returns
-------
dict
Mapping (ligand_atom_index, protein_atom_index) -> dict with key
'dists' initialized to an empty list. Distances over the trajectory
will be appended to this list by :func:`collect_distances`.
Behavior details
----------------
- Protein atoms are selected with the MDAnalysis "around" syntax.
- The returned atom indices are MDAnalysis global atom indices (0-based).
"""
lig_heavy = u.select_atoms(ligand_sel)
anchors = {}
for lig_atom in lig_heavy:
# select protein heavy atoms within cutoff of ligand atom
prot_sel = f"(protein or resname PRT) and (around {cutoff} index {lig_atom.index}) and (not name H*)"
prot_atoms = u.select_atoms(prot_sel)
for prot_atom in prot_atoms:
anchors[(lig_atom.index, prot_atom.index)] = {"dists": []}
return anchors
def collect_distances(u, anchors):
"""Populate `anchors` with distance time series from the trajectory.
Parameters
----------
u : MDAnalysis.Universe
Universe that has already been loaded with topology and trajectory.
anchors : dict
Dictionary produced by :func:`find_anchor_pairs`. Keys are (lig_idx, prot_idx).
Returns
-------
dict
The same anchors dict with 'dists' converted to numpy arrays and
augmented with 'avg_dist' and 'sd_dist' for each pair.
Notes
-----
- Distances are computed frame-by-frame using MDAnalysis distance routines
with periodic box handling (box=frame.dimensions).
- For performance, this function iterates over frames and pairs; for large
trajectories or many pairs consider vectorized approaches or sampling.
"""
# Iterate trajectory and collect distances for each pair
for frame in u.trajectory:
for lig_idx, prot_idx in list(anchors.keys()):
# compute distance using MDAnalysis distance routine
distance = dist(
mda.AtomGroup([u.atoms[lig_idx]]),
mda.AtomGroup([u.atoms[prot_idx]]),
box=frame.dimensions,
)[2][0]
anchors[(lig_idx, prot_idx)]["dists"].append(distance)
# convert lists to numpy arrays and compute stats
for pair in anchors.keys():
anchors[pair]["dists"] = np.array(anchors[pair]["dists"])
anchors[pair]["avg_dist"] = anchors[pair]["dists"].mean()
anchors[pair]["sd_dist"] = anchors[pair]["dists"].std()
return anchors
def report_low_sd_pairs(u, anchors, n=5):
"""Report and return the top-N anchor pairs with lowest distance SD.
Parameters
----------
u : MDAnalysis.Universe
Universe used only for pretty-printing atom information.
anchors : dict
Anchors dict returned by :func:`collect_distances`.
n : int
Number of pairs to report.
Returns
-------
list
Ordered list of (lig_idx, prot_idx) tuples sorted by ascending SD.
This function logs informative lines that can be included in a tutorial
to show how candidate anchor pairs are selected based on stability.
"""
ordered = [item[0] for item in sorted(anchors.items(), key=lambda it: it[1]["sd_dist"]) ]
LOG.info("Top %d pairs by lowest SD:", n)
for i, pair in enumerate(ordered[:n]):
a, b = pair
LOG.info("Pair %d: %s - avg %.2f SD %.2f", i + 1, (u.atoms[a], u.atoms[b]), anchors[pair]["avg_dist"], anchors[pair]["sd_dist"])
return ordered
def get_anchor_ats(a1_idx, u):
"""Select three anchor atoms around a given atom index.
Parameters
----------
a1_idx : int
Index of the primary anchor atom (0-based, MDAnalysis indexing).
u : MDAnalysis.Universe
Universe containing topology information - used to query bonded atoms.
Returns
-------
tuple
(a1_idx, a2_idx, a3_idx) where a2 and a3 are heavy atoms chosen to be
bonded neighbors of a1 (or the next atoms along the chain if at a terminus).
Behavior and edge cases
-----------------------
- The function prefers bonded heavy atoms that are not hydrogens.
- If a1 is terminal (only a single heavy neighbor), the function walks
one more bond along that neighbor to find a3.
- If no bonded heavy atoms are found an exception is raised; calling
code should ensure anchor candidates are sensible (heavy atoms typically have bonded neighbors).
Rationale
---------
These three anchor atoms are used to define internal angles/dihedrals
in Boresch-style restraints: the ligand anchor (a1,a2,a3) will be paired
with a receptor anchor triplet to define distance/angle/dihedral restraints.
"""
a1_at = u.atoms[a1_idx]
# select bonded heavy atoms (not hydrogens)
bonded_heavy = a1_at.bonded_atoms.select_atoms("not name H*")
if len(bonded_heavy) == 0:
raise RuntimeError(f"No bonded heavy atoms found for atom index {a1_idx}")
a2_idx = bonded_heavy[0].index
if len(bonded_heavy) > 1:
a3_idx = bonded_heavy[1].index
else:
# step one further away along heavy atoms
next_heavy = bonded_heavy[0].bonded_atoms.select_atoms("not name H*")
# prefer the second atom if that avoids going back to a1_idx
a3_candidate = next_heavy[1].index if len(next_heavy) > 1 else next_heavy[0].index
if a3_candidate == a1_idx and len(next_heavy) > 1:
a3_candidate = next_heavy[0].index
a3_idx = a3_candidate
return a1_idx, a2_idx, a3_idx
# Geometry helpers
def get_distance(idx1, idx2, u):
"""Compute Euclidean distance (Å) between two atoms by index.
Parameters
----------
idx1, idx2 : int
Atom indices (0-based).
u : MDAnalysis.Universe
Returns
-------
float
Distance in Å.
"""
return dist(mda.AtomGroup([u.atoms[idx1]]), mda.AtomGroup([u.atoms[idx2]]), box=u.dimensions)[2][0]
def get_angle(idx1, idx2, idx3, u):
"""Compute angle (radians) defined by three atoms A-B-C.
Parameters
----------
idx1 : int
Index of atom C (first argument in original function definition)
idx2 : int
Index of atom B (central atom)
idx3 : int
Index of atom A (last argument)
u : MDAnalysis.Universe
Returns
-------
float
Angle in radians.
Notes
-----
The ordering follows the original script: get_angle(idx1, idx2, idx3)
returns the angle at atom idx2 formed by vectors (A-B) and (C-B) where
A = idx3, B = idx2, C = idx1.
"""
C = u.atoms[idx1].position
B = u.atoms[idx2].position
A = u.atoms[idx3].position
BA = A - B
BC = C - B
angle = np.arccos(np.dot(BA, BC) / (norm(BA) * norm(BC)))
return angle
def get_dihedral(idx1, idx2, idx3, idx4, u):
"""Compute dihedral (torsion) angle in radians for four atoms.
Parameters
----------
idx1, idx2, idx3, idx4 : int
Atom indices specifying the torsion in the order used by MDAnalysis.
u : MDAnalysis.Universe
Returns
-------
float
Dihedral angle in radians (range -pi..pi).
Notes
-----
MDAnalysis' calc_dihedrals returns values in the range (-pi, pi].
"""
positions = [u.atoms[idx].position for idx in [idx1, idx2, idx3, idx4]]
dihedral = calc_dihedrals(positions[0], positions[1], positions[2], positions[3], box=u.dimensions)
return dihedral
def get_boresch_dof(l1, l2, l3, r1, r2, r3, u):
"""Compute the Boresch degrees of freedom for a given anchor sextet.
Parameters
----------
l1,l2,l3 : int
Indices of the three ligand anchor atoms (l1 is bonded to receptor r1).
r1,r2,r3 : int
Indices of the three receptor anchor atoms (r1 bonded to ligand l1).
u : MDAnalysis.Universe
Returns
-------
tuple
(r, thetaA, thetaB, phiA, phiB, phiC, thetaR, thetaL)
Where
-----
- r : float, distance between r1 and l1 (Å)
- thetaA, thetaB : floats, angles (radians)
- phiA, phiB, phiC : floats, dihedrals (radians)
- thetaR, thetaL : internal receptor/ligand angles (radians)
Usage
-----
These values correspond to the common variables used to define a set
of Boresch restraints (one distance, two angles, three dihedrals).
"""
# Ordering r3,r2,r1,l1,l2,l3 as in original
r = get_distance(r1, l1, u)
thetaA = get_angle(r2, r1, l1, u)
thetaB = get_angle(r1, l1, l2, u)
phiA = get_dihedral(r3, r2, r1, l1, u)
phiB = get_dihedral(r2, r1, l1, l2, u)
phiC = get_dihedral(r1, l1, l2, l3, u)
thetaR = get_angle(r3, r2, r1, u)
thetaL = get_angle(l1, l2, l3, u)
return r, thetaA, thetaB, phiA, phiB, phiC, thetaR, thetaL
def analyze_boresch_over_traj(u, pairs, max_pairs=200):
"""Compute time series and statistics for Boresch DOF across trajectory.
Parameters
----------
u : MDAnalysis.Universe
Universe with trajectory.
pairs : iterable
Iterable of (l1_idx, r1_idx) anchor pairs to analyze.
max_pairs : int
Maximum number of pairs to process (for performance control).
Returns
-------
dict
Mapping pair -> dict with keys:
- 'anchor_ats': [l1,l2,l3,r1,r2,r3]
- for each dof e.g. 'r', 'phiA', etc.: dict with 'values' (np.array), 'avg', 'sd', 'k'
- 'tot_var': sum of variances used to rank pair suitability
Important details
-----------------
- For dihedral DOFs (phiA/B/C) this function applies a circular-statistics
aware procedure to compute mean and standard deviation so that periodicity
is correctly handled (angles near -pi and +pi are treated continuously).
- Force constants k are estimated assuming a Gaussian distribution via
k = RT / sd^2, where RT is approximated as 0.593 kcal/mol at 289 K in
the original script. This is the same convention and will be halved
later when writing restraint parameters to be consistent with external tools.
"""
boresch = {}
boresch_dof_list = ["r", "thetaA", "thetaB", "phiA", "phiB", "phiC", "thetaR", "thetaL"]
for pair in pairs[:max_pairs]:
boresch[pair] = {}
l1_idx, r1_idx = pair
_, l2_idx, l3_idx = get_anchor_ats(l1_idx, u)
_, r2_idx, r3_idx = get_anchor_ats(r1_idx, u)
boresch[pair]["anchor_ats"] = [l1_idx, l2_idx, l3_idx, r1_idx, r2_idx, r3_idx]
for dof in boresch_dof_list:
boresch[pair][dof] = {"values": []}
n_frames = len(u.trajectory)
for i, frame in enumerate(u.trajectory):
r, thetaA, thetaB, phiA, phiB, phiC, thetaR, thetaL = get_boresch_dof(
l1_idx, l2_idx, l3_idx, r1_idx, r2_idx, r3_idx, u
)
boresch[pair]["r"]["values"].append(r)
boresch[pair]["thetaA"]["values"].append(thetaA)
boresch[pair]["thetaB"]["values"].append(thetaB)
boresch[pair]["phiA"]["values"].append(phiA)
boresch[pair]["phiB"]["values"].append(phiB)
boresch[pair]["phiC"]["values"].append(phiC)
boresch[pair]["thetaR"]["values"].append(thetaR)
boresch[pair]["thetaL"]["values"].append(thetaL)
# After frames loop compute statistics
boresch[pair]["tot_var"] = 0.0
for dof in boresch_dof_list:
values = np.array(boresch[pair][dof]["values"])
boresch[pair][dof]["values"] = values
avg = values.mean()
boresch[pair][dof]["avg"] = avg
if dof.startswith("phi"):
# circular statistics handling for dihedrals
corrected_vals = []
for val in values:
dtheta = abs(val - avg)
corrected_vals.append(min(dtheta, 2 * np.pi - dtheta))
corrected_vals = np.array(corrected_vals)
boresch[pair][dof]["sd"] = corrected_vals.std()
# correct mean
periodic_bound = avg - np.pi
if periodic_bound < -np.pi:
periodic_bound += 2 * np.pi
corrected_avg_vals = [v + 2 * np.pi if v < periodic_bound else v for v in values]
m_corr = np.array(corrected_avg_vals).mean()
if m_corr > np.pi:
boresch[pair][dof]["avg"] = m_corr - 2 * np.pi
else:
boresch[pair][dof]["avg"] = m_corr
else:
boresch[pair][dof]["sd"] = values.std()
if dof not in ("thetaR", "thetaL"):
boresch[pair]["tot_var"] += boresch[pair][dof]["sd"] ** 2
boresch[pair][dof]["k"] = 0.593 / (boresch[pair][dof]["sd"] ** 2)
return boresch
def select_pairs(boresch_dict):
"""Select suitable anchor pairs based on geometric filters.
Parameters
----------
boresch_dict : dict
Output from :func:`analyze_boresch_over_traj`.
Returns
-------
list
Filtered list of pairs, ordered by increasing tot_var.
Filtering criteria
------------------
- Distance r average must be > 1.0 Å (filters out extremely close/invalid pairs)
- Average angles thetaA and thetaB must be within (0.52, 2.62) radians (~30°-150°)
These thresholds were chosen empirically to avoid degenerate restraints.
"""
# order by tot_var
ordered = [item[0] for item in sorted(boresch_dict.items(), key=lambda it: it[1]["tot_var"]) ]
# filter by distance and internal angles
selected = []
for pair in ordered:
cond_dist = boresch_dict[pair]["r"]["avg"] > 1.0
avg_angles = [boresch_dict[pair][a]["avg"] for a in ("thetaA", "thetaB")]
cond_angles = all((0.52 < ang < 2.62) for ang in avg_angles)
if cond_dist and cond_angles:
selected.append(pair)
return selected
def plot_time_series(boresch_dict, pairs, fig_prefix="boresch", num_pairs=5):
"""Plot time series for each Boresch DOF for the top candidate pairs.
The plotting function will unwrap phi (dihedral) time series around the
computed circular mean so that the plot does not show large jumps at the
-π/π periodic boundary. Figures are saved with the prefix provided.
Parameters
----------
boresch_dict : dict
Boresch results as produced by :func:`analyze_boresch_over_traj`.
pairs : list
Candidate pairs (ordering is preserved); only the first `num_pairs`
are plotted.
fig_prefix : str
Prefix used for saved figure filenames.
num_pairs : int
Number of pairs to include in the plot (default 5).
"""
dofs = ["r", "thetaA", "thetaB", "phiA", "phiB", "phiC", "thetaR", "thetaL"]
n_dof = len(dofs)
fig, axs = plt.subplots(1, n_dof, figsize=(2.6 * n_dof, 6))
def _unwrap_to_mean(values, mean):
"""Shift angular values by multiples of 2*pi so they lie within (mean-pi, mean+pi]."""
return mean + ((values - mean + np.pi) % (2 * np.pi) - np.pi)
for i, dof in enumerate(dofs):
for j, pair in enumerate(list(boresch_dict.keys())[:num_pairs]):
vals = boresch_dict[pair][dof]["values"]
# For dihedrals (phi) unwrap values around their mean to avoid jumps at the -pi/pi boundary
if dof.startswith("phi"):
mean = boresch_dict[pair][dof].get("avg", 0.0)
vals_plot = _unwrap_to_mean(vals, mean)
else:
vals_plot = vals
axs[i].plot(np.arange(len(vals_plot)), vals_plot, label=f"Pair {pair}")
axs[i].set_xlabel("Frame No")
axs[i].set_ylabel("r (\u212B)" if dof == "r" else f"{dof} (rad)")
axs[i].legend()
fig.tight_layout()
fig_file = f"{fig_prefix}_timeseries.png"
fig.savefig(fig_file)
LOG.info("Saved time series figure to %s", fig_file)
def plot_histograms(boresch_dict, pairs, fig_prefix="boresch", highlight_pairs=None):
"""Plot histograms of DOF distributions for top candidate pairs.
Parameters
----------
boresch_dict : dict
Boresch data structure.
pairs : list
Candidate pairs selected for plotting.
fig_prefix : str
Prefix for saved histogram images.
Notes
-----
- Histograms include a vertical dashed line showing the computed average.
- Dihedral values are plotted in their stored range (after circular
averaging), which avoids misleading binning across the -π/π boundary.
"""
chosen = pairs[:3]
dof_order = ["r", "thetaA", "thetaB", "phiA", "phiB", "phiC"]
fig, axs = plt.subplots(len(chosen), len(dof_order), figsize=(16, 4 * len(chosen)))
for pi, pair in enumerate(chosen):
for di, dof in enumerate(dof_order):
ax = axs[pi][di] if len(chosen) > 1 else axs[di]
vals = boresch_dict[pair][dof]["values"]
# draw histogram and get patch artists so we can outline selected pairs
n, bins, patches = ax.hist(vals, bins=10)
ax.axvline(x=boresch_dict[pair][dof]["avg"], color="r", linestyle="dashed", linewidth=2)
# If this pair is in the highlight set, outline the bars and add title
if highlight_pairs and pair in highlight_pairs:
for p in patches:
p.set_edgecolor('red')
p.set_linewidth(1.2)
p.set_alpha(0.6)
ax.set_title(f"Selected pair: {pair}")
ax.set_xlabel("r (\u212B)" if dof == "r" else f"{dof} (rad)")
ax.set_ylabel("Num Vals")
fig.tight_layout()
fig_file = f"{fig_prefix}_histograms.png"
fig.savefig(fig_file)
LOG.info("Saved histogram figure to %s", fig_file)
def print_boresch_params(pair, boresch_dict, out_rest="rest.in"):
"""Write out restraint parameters for a chosen anchor pair.
Parameters
----------
pair : tuple
Anchor pair tuple returned by selection routines.
boresch_dict : dict
Results structure from :func:`analyze_boresch_over_traj`.
out_rest : str
Output filename for the AMBER restraint file (default 'rest.in').
Behavior
--------
- Prints a JSON-like single-line summary to stdout suitable for parsing
by tutorial readers or downstream tools.
- Writes an AMBER &rst formatted restraint file where atom indices are
converted to AMBER's 1-based indexing.
- Force constants are halved in the output to match the energy definition
used by some external MD codes (consistent with the original script).
"""
l1, l2, l3, r1, r2, r3 = boresch_dict[pair]["anchor_ats"]
r0 = boresch_dict[pair]["r"]["avg"]
thetaA0 = boresch_dict[pair]["thetaA"]["avg"]
thetaB0 = boresch_dict[pair]["thetaB"]["avg"]
phiA0 = boresch_dict[pair]["phiA"]["avg"]
phiB0 = boresch_dict[pair]["phiB"]["avg"]
phiC0 = boresch_dict[pair]["phiC"]["avg"]
kr = boresch_dict[pair]["r"]["k"] / 2
kthetaA = boresch_dict[pair]["thetaA"]["k"] / 2
kthetaB = boresch_dict[pair]["thetaB"]["k"] / 2
kphiA = boresch_dict[pair]["phiA"]["k"] / 2
kphiB = boresch_dict[pair]["phiB"]["k"] / 2
kphiC = boresch_dict[pair]["phiC"]["k"] / 2
json_str = (
'{"anchor_points":{"r1":%d, "r2":%d, "r3":%d, "l1":%d, "l2":%d, "l3":%d},'
'"equilibrium_values":{"r0":%.2f, "thetaA0":%.2f, "thetaB0":%.2f, "phiA0":%.2f, "phiB0":%.2f, "phiC0":%.2f},'
'"force_constants":{"kr":%.2f, "kthetaA":%.2f, "kthetaB":%.2f, "kphiA":%.2f, "kphiB":%.2f, "kphiC":%.2f}}'
) % (r1, r2, r3, l1, l2, l3, r0, thetaA0, thetaB0, phiA0, phiB0, phiC0, kr, kthetaA, kthetaB, kphiA, kphiB, kphiC)
print(json_str)
# write AMBER restraint input (indices are 1-based in AMBER)
with open(out_rest, "w") as fh:
fh.write(f"&rst iat={r1+1},{l1+1},0\n")
fh.write(f" r1={0.0:.5f},r2={r0:.5f},r3={r0:.5f},r4=999.000,rk2={kr:.2f}, rk3={kr:.2f}/\n")
fh.write(f"&rst iat={r2+1},{r1+1},{l1+1},0\n")
fh.write(f" r1={-180.0:.5f},r2={np.degrees(thetaA0):.5f},r3={np.degrees(thetaA0):.5f},r4=180.000,rk2={kthetaA:.2f}, rk3={kthetaA:.2f}/\n")
fh.write(f"&rst iat={r1+1},{l1+1},{l2+1},0\n")
fh.write(f" r1={-180.0:.5f},r2={np.degrees(thetaB0):.5f},r3={np.degrees(thetaB0):.5f},r4=180.000,rk2={kthetaB:.2f}, rk3={kthetaB:.2f}/\n")
fh.write(f"&rst iat={r3+1},{r2+1},{r1+1},{l1+1},0\n")
fh.write(f" r1={-180.0:.5f},r2={np.degrees(phiA0):.5f},r3={np.degrees(phiA0):.5f},r4=180.000,rk2={kphiA:.2f}, rk3={kphiA:.2f}/\n")
fh.write(f"&rst iat={r2+1},{r1+1},{l1+1},{l2+1},0\n")
fh.write(f" r1={-180.0:.5f},r2={np.degrees(phiB0):.5f},r3={np.degrees(phiB0):.5f},r4=180.000,rk2={kphiB:.2f}, rk3={kphiB:.2f}/\n")
fh.write(f"&rst iat={r1+1},{l1+1},{l2+1},{l3+1},0\n")
fh.write(f" r1={-180.0:.5f},r2={np.degrees(phiC0):.5f},r3={np.degrees(phiC0):.5f},r4=180.000,rk2={kphiC:.2f}, rk3={kphiC:.2f}/\n")
LOG.info("Wrote restraint file %s", out_rest)
def main():
"""Main driver for the script.
Loads the universe, identifies candidate anchor pairs, computes statistics,
selects suitable pairs, writes restraint input, and saves diagnostic figures.
This function orchestrates the overall workflow and emits informative
logging messages for tutorial users to follow along.
"""
args = parse_args()
logging.basicConfig(level=logging.INFO, format="%(asctime)s - %(levelname)s - %(message)s")
u = mda.Universe(args.topology, args.trajectory)
LOG.info("Loaded universe: %s, trajectory frames: %d", args.topology, len(u.trajectory))
anchors = find_anchor_pairs(u, ligand_sel=args.ligand_sel, cutoff=args.cutoff)
if not anchors:
LOG.error("No candidate anchors found. Adjust ligand selection or cutoff.")
return
anchors = collect_distances(u, anchors)
ordered_pairs = report_low_sd_pairs(u, anchors, n=args.num_pairs)
# Analyze boresch degrees of freedom over trajectory for top pairs
boresch = analyze_boresch_over_traj(u, ordered_pairs, max_pairs=args.max_pairs_traj)
# Order by variance and select suitable pairs
pairs_by_var = [item[0] for item in sorted(boresch.items(), key=lambda it: it[1]["tot_var"]) ]
LOG.info("Pairs ordered by tot_var (top 10): %s", pairs_by_var[:10])
selected = select_pairs(boresch)
LOG.info("Selected pairs after filtering: %s", selected)
if selected:
# plot and write restraints for the first selected pair
plot_time_series(boresch, selected, fig_prefix=args.fig_prefix, num_pairs=args.num_pairs)
plot_histograms(boresch, selected, fig_prefix=args.fig_prefix, highlight_pairs=selected)
print_boresch_params(selected[0], boresch, out_rest=args.out_rest)
else:
LOG.warning("No suitable pairs found after filtering. Increase candidates or relax filters.")
if __name__ == "__main__":
main()
This code uses the MDAnalysis package to analyze the trajectory and select appropriate atoms for the Boresch restraints.
You can run the code with the following command:
python generate_boresch_restraints.py
This will create a couple of image files, as well as a “rest.in” file that contains the necessary restraint information for use in an Amber simulation.
There are two image files that are generated by the script.
The first is boresch_histograms.png, which shows the histograms of the distance, angle, and dihedral values for the selected atoms over the course of the trajectory. This is useful for visualizing how well the selected atoms maintain their relative positions and orientations.
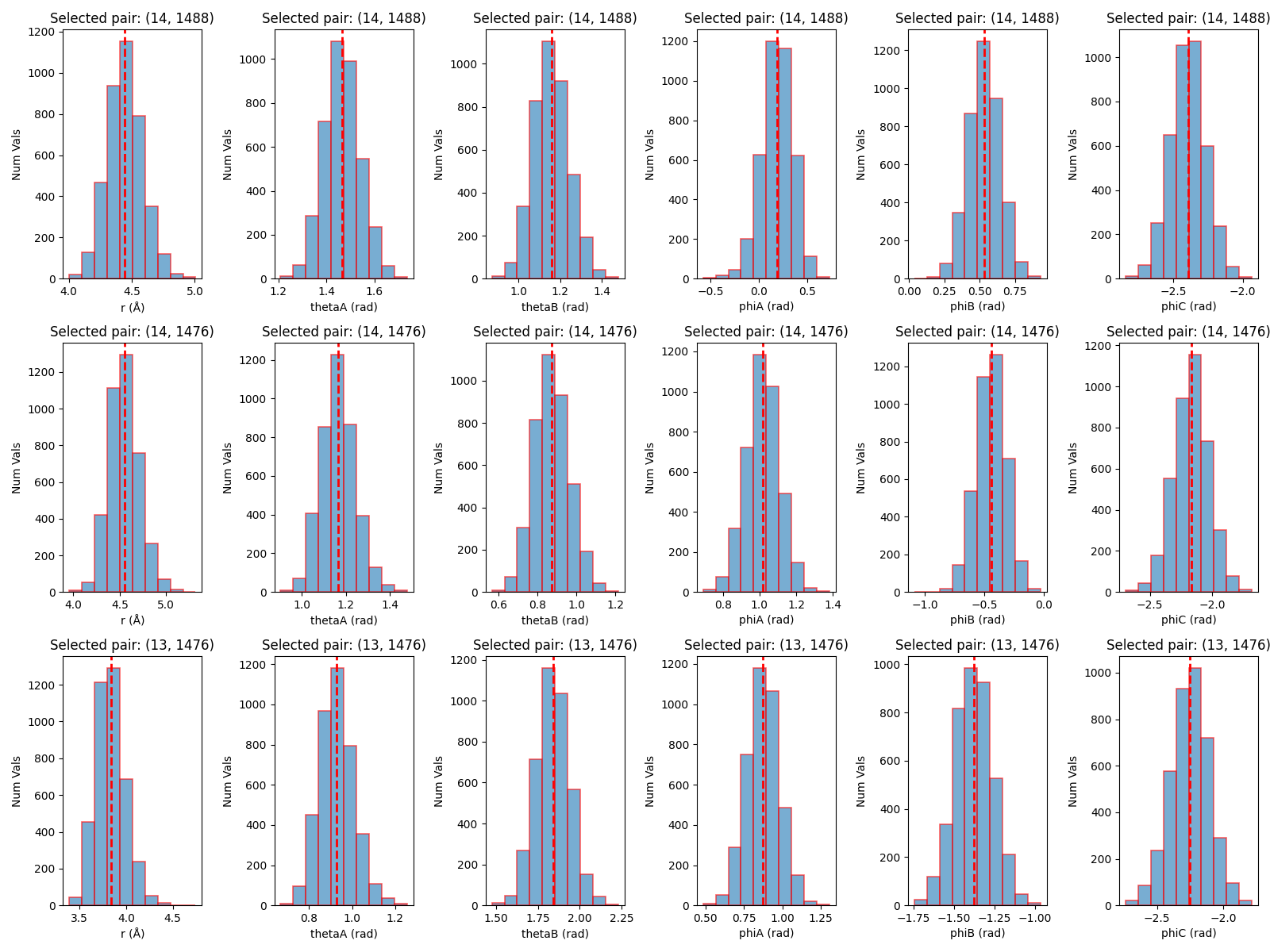
The second image is boresch_timeseries.png, which shows the time series of the distance, angle, and dihedral values for the selected atoms over the course of the trajectory. This is useful for visualizing how stable the selected atoms are over time.
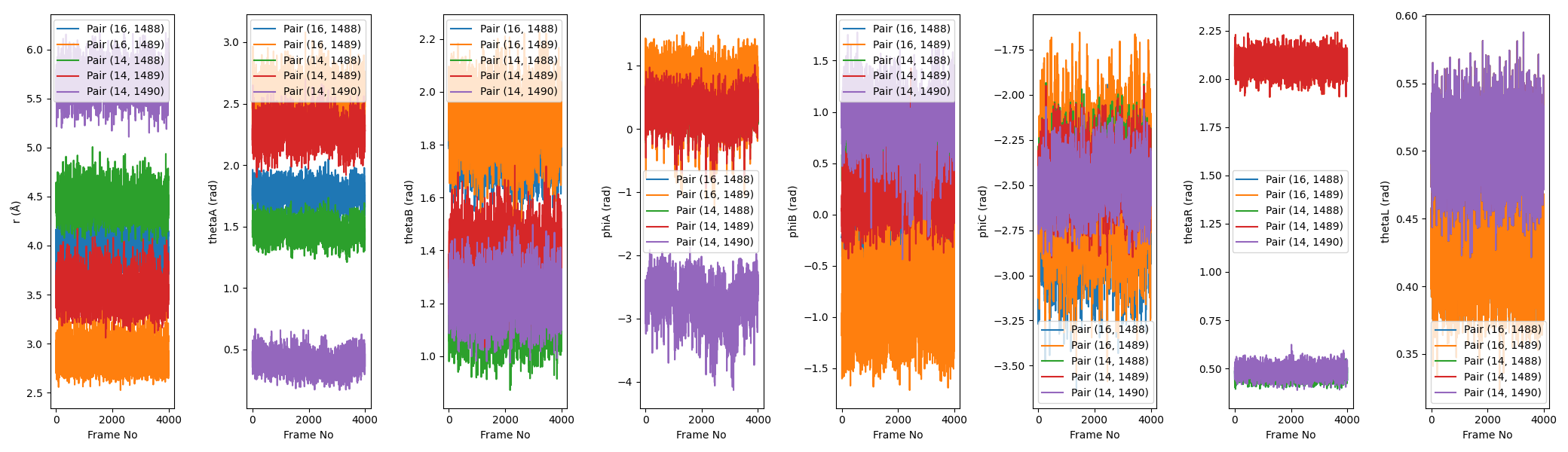
The final output is the rest.in file, which contains the necessary restraint information for use in an Amber simulation. This file includes the atom indices, equilibrium values, and force constants for the Boresch restraints.
An example of the rest.in file generated is shown below:
&rst iat=1489,15,0
r1=0.00000,r2=4.44575,r3=4.44575,r4=999.000,rk2=15.06, rk3=15.06/
&rst iat=1477,1489,15,0
r1=-180.00000,r2=83.99734,r3=83.99734,r4=180.000,rk2=53.21, rk3=53.21/
&rst iat=1489,15,14,0
r1=-180.00000,r2=66.38281,r3=66.38281,r4=180.000,rk2=40.55, rk3=40.55/
&rst iat=1490,1477,1489,15,0
r1=-180.00000,r2=11.03062,r3=11.03062,r4=180.000,rk2=29.83, rk3=29.83/
&rst iat=1477,1489,15,14,0
r1=-180.00000,r2=30.50819,r3=30.50819,r4=180.000,rk2=62.83, rk3=62.83/
&rst iat=1489,15,14,16,0
r1=-180.00000,r2=-137.04995,r3=-137.04995,r4=180.000,rk2=49.20, rk3=49.20/
Tip
The rest.in file should look familiar to you if you are familiar with Amber’s NMR restraint format, as Boresch restraints are implemented using the same underlying machinery.
Specifically, each &rst block in the rest.in file corresponds to a specific restraint type (distance, angle, dihedral) and contains the necessary parameters for that restraint.
You can tell what type of restraint it is based on the number of atoms involved:
Distance restraints involve 2 atoms
Angle restraints involve 3 atoms
Dihedral restraints involve 4 atoms
So the first line in rest.in is a distance restraint, since iat=1489,15
Now, the line after the &rst marker specifies the restraint values. Specifically, the fields always are:
r1: the lower bound where the restraint should reach infinity
r2: a mid-point of the restraint well where it should start taking effect (on its way to r1)
r3: another mid-point of the restraint well where it should start taking effect (on its way to r4)
r4: the upper bound where the restraint should reach infinity
rk2: the force constant for the left side of the well (between r1 and r2)
rk3: the force constant for the right side of the well (between r3 and r4)
Note
In between r2 and r3, there is no force applied (though, here you’ll note for all restraints r2=r3, meaning the well is harmonic in all cases.)
With these boresch restraints generated, you are now ready to use them in your ABFE calculations!
Setting Up and Running Annihilation Simulations
For this tutorial, we will need two things to get started. The first is the equilibrated end-state files from the previous tutorial:ref:Equilibrating the End States of Tyk2 for ABFE Calculations <equilibrate_endstates>; and the second is the Boresch restraint files generated in the tutorial:ref:Generating Boresch Restraints for Tyk2 ABFE Calculations <boresch_restraints>. For your convenience, we have provided the necessary files below to get you started.
Attention
Update the download link to point to the finalized tutorial files once they are uploaded.
Choosing A Lambda Schedule
The first step in setting up an ABFE calculation is to define a lambda schedule. A later tutorial will cover this topic in more detail with information on how to optimize lambda schedules for your specific system; however, for the purposes of this tutorial we will use a linear lambda schdule with 11 windows.
Warning
A linear lambda schedule is usually not optimizal for ABFE calculations, and ABFE calculations often need far more than 11 windows to converge. This tutorial is meant to demonstrate the basic steps of setting up and running an ABFE calculation, and is not intended to produce converged or realistic results.
The lambda schedule we will use is as follows:
Window Lambda Value
---------------------
0 0.0
1 0.1
2 0.2
3 0.3
4 0.4
5 0.5
6 0.6
7 0.7
8 0.8
9 0.9
10 1.0
Annihilation Simulation Setup
With a lambda schedule in place, we can now slowly annihilate the ligand from the equilibrated complex and binder end-states. Roughly, in both cases this involves starting from the end-state where \(\lambda = 0\) and gradually turning off the ligand’s interactions with its environment until it is fully decoupled at \(\lambda = 1\).
&cntrl
imin = 1,
maxcyc = 5000,
ncyc = 100,
ntx = 1,
ntmin = 2,
ntwe = 0,
ntwr = 0,
ntpr = 1000,
ntb = 1,
ntp = 0,
cut = 10.0,
ntr = 0,
ioutfm=1,
ntxo=1,
ifsc = 1
icfe = 1
clambda = REPLACE_WITH_LAMBDA
timask1 = ':1'
timask2 = ''
scmask1 = ':1'
scmask2 = ''
scalpha = 0.5
scbeta = 1.0
gti_cut = 1
gti_output = 1
gti_add_sc = 25
gti_scale_beta = 1
gti_cut_sc_on = 8
gti_cut_sc_off = 10
gti_lam_sch = 1
gti_ele_sc = 1
gti_vdw_sc = 1
gti_cut_sc = 2
gti_ele_exp = 2
gti_vdw_exp = 2
gti_syn_mass = 0
gremd_acyc = 1
gti_bat_sc = -1
/
&cntrl
imin = 1,
maxcyc = 5000,
ncyc = 100,
ntx = 1,
ntmin = 2,
ntwe = 0,
ntwr = 0,
ntpr = 1000,
ntb = 1,
ntp = 0,
cut = 10.0,
ntr = 0,
ioutfm=1,
ntxo=1,
ifsc = 1
icfe = 1
clambda = REPLACE_WITH_LAMBDA
timask1 = ':1'
timask2 = ''
scmask1 = ':1'
scmask2 = ''
scalpha = 0.5
scbeta = 1.0
gti_cut = 1
gti_output = 1
gti_add_sc = 25
gti_scale_beta = 1
gti_cut_sc_on = 8
gti_cut_sc_off = 10
gti_lam_sch = 1
gti_ele_sc = 1
gti_vdw_sc = 1
gti_cut_sc = 2
gti_ele_exp = 2
gti_vdw_exp = 2
gti_syn_mass = 0
gremd_acyc = 1
gti_bat_sc = -1
nmropt = 1
/
&wt type = 'END', /
DISANG=rest.in
/
Above, we have included the mdin files used to annihilate the ligand in both the binder and the complex environments. Note. These mdin files contain a placeholder for the specific lambda value for each window. This is REPLACE_WITH_LAMBDA.
We will start by generating the lambda windows for both the binder and complex annihilation simulations. To do this, we will start with our simple lambda schedule defined above.
Start by creating a working_directory for these steps within the abfe_files directory. We will make separate directories for the binder and complex annihilation simulations within this folder.
mkdir working_dir
cd working_dir
mkdir binder complex
Next, we will use a bash for loop to generate the mdin files for each lambda window for both the binder and complex annihilation simulations.
# Binder Annhilation Windows
lambda_values=(0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0)
for i in ${!lambda_values[@]}
do
lambda_value=${lambda_values[$i]}
sed "s/REPLACE_WITH_LAMBDA/$lambda_value/g" ../binder_mdins/annhilate.mdin > binder/annihilate_lambda_${i}.mdin
done
# Complex Annhilation Windows
for i in ${!lambda_values[@]}
do
lambda_value=${lambda_values[$i]}
sed "s/REPLACE_WITH_LAMBDA/$lambda_value/g" ../complex_mdins/annhilate.mdin > complex/annihilate_lambda_${i}.mdin
done
Now, you should have 11 mdin files for both the binder and complex annihilation simulations, each corresponding to a different lambda window.
Now, we can proceed to run these lambda windows iteratively starting from the \(\lambda = 0\) window and moving to the next window using the output of the previous window as input.
We will start with the binder simulations. Here, we will use the equilibrated end-state files (provided with the tutorial files) as input for the first window (\(\lambda = 0\)), and then use the output of each window as input for the next window.
cd binder
# Run Lambda Windows Iteratively
for i in {0..10}
do
if [ $i -eq 0 ]
then
# First window uses equilibrated end-state files
pmemd.cuda -O -i annihilate_lambda_${i}.mdin -p ../../topology/binder_ejm_31_hmr.parm7 -c ../../topology/binder_ejm_31_unrestrained.rst7 -o annihilate_binder_${i}.mdout -r annihilate_binder_${i}.rst7 -x annihilate_binder_${i}_traj.nc -ref ../../topology/binder_ejm_31_unrestrained.rst7
else
# Subsequent windows use output of previous window
pmemd.cuda -O -i annihilate_lambda_${i}.mdin -p ../../topology/binder_ejm_31_hmr.parm7 -c annihilate_binder_$((i-1)).rst7 -o annihilate_binder_${i}.mdout -r annihilate_binder_${i}.rst7 -x annihilate_binder_${i}_traj.nc -ref annihilate_binder_$((i-1)).rst7
fi
done
These commands will run the binder annilation simulations for each lambda window iteratively. These are minimizations, and thus are relatively quick to run.
For the complex simulations, we will follow the same procedure, except we also need to include the Boresch restraint rest.in file, as well as the lambda.sch file that defines how the restraints are scaled during the annihilation process.
This lambda.sch file controls how the Boresch restraints are scaled during the annihilation process to ensure that the ligand remains properly restrained as its interactions with the environment are turned off.
TypeRestBA, smooth_step2, symmetric, 1.0, 0.0
Warning
The lambda.sch is essential for the proper scaling of Boresch restraints during the annihilation process. If not included, the restraints will be applied to the real state and then slowly turned off, which is exactly the opposite of what we want to achieve. This is a very common mistake when setting up ABFE calculations with Boresch restraints, and will lead to incorrect results.
Thus, we will copy these files into the complex working directory before running the simulations.
cd ../complex
cp ../../boresch_restraints/rest.in .
cp ../../boresch_restraints/lambda.sch .
# Run Lambda Windows Iteratively
for i in {0..10}
do
if [ $i -eq 0 ]
then
# First window uses equilibrated end-state files
pmemd.cuda -O -i annihilate_lambda_${i}.mdin -p ../../topology/target_ejm_31_hmr.parm7 -c ../../topology/complex_ejm_31_unrestrained.rst7 -o annihilate_complex_${i}_out.log -r annihilate_complex_${i}.rst7 -x annihilate_complex_${i}.nc -ref ../../topology/complex_ejm_31_unrestrained.rst7
else
# Subsequent windows use output of previous window
pmemd.cuda -O -i annihilate_lambda_${i}.mdin -p ../../topology/target_ejm_31_hmr.parm7 -c annihilate_complex_$((i-1)).rst7 -o annihilate_complex_${i}.mdout -r annihilate_complex_${i}.rst7 -x annihilate_complex_${i}.nc -ref annihilate_complex_$((i-1)).rst7
fi
done
These calculations will run relatively quickly, as they are minimizations. After this, we can move on to equilibrating the lambda windows.
Equilibrating the Lambda Windows
We will now equilibrate each of the lambda windows generated in the previous step. Unlike the annihilation simulations, we will run the equilibration simulations for each lambda window in parallel, as they are independent of each other. Currently, these parallel runs will not be using replica exchange - replica exchange won’t turn on until the production phase.
Binder Equilibration
We will again start with the binder, equilibrating each lambda window using the corresponding mdin files provided below.
.
&cntrl
imin = 0
nstlim = 100000
dt = 0.001
irest = 0
ntx = 1
ntxo = 1
ntc = 2
ntf = 1
ntwx = 0
ntwr = 0
ntpr = 125
cut = 10.0
iwrap = 0
ntb = 1
ntp = 0
tempi = 200
temp0 = 298
ntt = 3
gamma_ln = 5.0
barostat = 2
pres0 = 1.01325
taup = 1
rmsd_mask(1) = ':1 & !@H='
rmsd_strength(1)= 0
rmsd_ti(1) = 2
rmsd_type = 0
ig = -1
ifsc = 1
icfe = 1
clambda = REPLACE_WITH_LAMBDA
timask1 = ':1'
timask2 = ''
crgmask = ""
scmask1 = ':1'
scmask2 = ''
scalpha = 0.5
scbeta = 1.0
gti_cut = 1
gti_output = 1
gti_add_sc = 25
gti_scale_beta = 1
gti_cut_sc_on = 8
gti_cut_sc_off = 10
gti_lam_sch = 1
gti_ele_sc = 1
gti_vdw_sc = 1
gti_cut_sc = 2
gti_ele_exp = 2
gti_vdw_exp = 2
gti_syn_mass = 0
gremd_acyc = 1
/
&wt
TYPE='TEMP0',
ISTEP1=0, ISTEP2=100000,
VALUE1=200, VALUE2=298,
&cntrl
imin = 0
nstlim = 50000
dt = 0.002
irest = 1
ntx = 5
ntxo = 1
ntc = 2
ntf = 1
ntwx = 0
ntwr = 0
ntpr = 125
cut = 10.0
iwrap = 0
ntb = 1
ntp = 0
tempi = 298
temp0 = 298
ntt = 3
gamma_ln = 5.0
barostat = 2
pres0 = 1.01325
taup = 1
ig = -1
ifsc = 1
icfe = 1
rmsd_mask(1) = ':1 & !@H='
rmsd_strength(1)= 0
rmsd_ti(1) = 2
rmsd_type = 0
clambda = REPLACE_WITH_LAMBDA
timask1 = ':1'
timask2 = ''
crgmask = ""
scmask1 = ':1'
scmask2 = ''
scalpha = 0.5
scbeta = 1.0
gti_cut = 1
gti_output = 1
gti_add_sc = 25
gti_scale_beta = 1
gti_cut_sc_on = 8
gti_cut_sc_off = 10
gti_lam_sch = 1
gti_ele_sc = 1
gti_vdw_sc = 1
gti_cut_sc = 2
gti_ele_exp = 2
gti_vdw_exp = 2
gti_syn_mass = 0
gremd_acyc = 1
/
&cntrl
imin = 0
nstlim = 250000
dt = 0.004
irest = 1
ntx = 5
ntxo = 1
ntc = 2
ntf = 1
ntwx = 0
ntwr = 0
ntpr = 125
cut = 10.0
iwrap = 0
ntb = 1
ntp = 0
tempi = 298
temp0 = 298
ntt = 3
gamma_ln = 5.0
barostat = 2
pres0 = 1.01325
taup = 1
ig = -1
ifsc = 1
icfe = 1
rmsd_mask(1) = ':1 & !@H='
rmsd_strength(1)= 0
rmsd_ti(1) = 2
rmsd_type = 0
clambda = REPLACE_WITH_LAMBDA
timask1 = ':1'
timask2 = ''
crgmask = ""
scmask1 = ':1'
scmask2 = ''
scalpha = 0.5
scbeta = 1.0
gti_cut = 1
gti_output = 1
gti_add_sc = 25
gti_scale_beta = 1
gti_cut_sc_on = 8
gti_cut_sc_off = 10
gti_lam_sch = 1
gti_ele_sc = 1
gti_vdw_sc = 1
gti_cut_sc = 2
gti_ele_exp = 2
gti_vdw_exp = 2
gti_syn_mass = 0
gremd_acyc = 1
/
Tip
These equilibration mdin files contain a placeholder for the specific lambda value for each window. This is REPLACE_WITH_LAMBDA.
Warning
Remember that mdin files REQUIRE a blank line at the end of the file to function properly with Amber. If you get an error that there was an unexpected end of file while reading the mdin, check to make sure there is a blank line at the end of your mdin file. If you use the provided mdin files, they already have this blank line.
Just like before, we will generate the mdins programmatically for each lambda window using a bash for loop.
cd ../binder
# Generate Equilibration MDINs for Each Lambda Window
lambda_values=(0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0)
for i in {0..10}
do
lambda_value=${lambda_values[$i]}
sed "s/REPLACE_WITH_LAMBDA/$lambda_value/g" ../../binder_mdins/heat.mdin > heat_lambda_${i}.mdin
sed "s/REPLACE_WITH_LAMBDA/$lambda_value/g" ../../binder_mdins/equil1.mdin > equil1_lambda_${i}.mdin
sed "s/REPLACE_WITH_LAMBDA/$lambda_value/g" ../../binder_mdins/equil2.mdin > equil2_lambda_${i}.mdin
done
Now, we also need a groupfile for the replica exchange simulations. This file will look a lot like the pmemd.CUDA command calls that we made before, except the groupfile will contain all of the lambda windows to be run in parallel.
The script below will generate the groupfile for the binder equilibration simulations.
rm binder_heat_groupfile.in binder_equil1_groupfile.in binder_equil2_groupfile.in
for i in {0..10}
do
echo "-O -i heat_${i}.mdin -p ../../topology/binder_ejm_31_hmr.parm7 -c annihilate_binder_${i}.rst7 -o binder_heat_${i}.mdout -r binder_heat_${i}.rst7 -x binder_heat_${i}.nc -ref ../../topology/binder_ejm_31_unrestrained.rst7" >> binder_heat_groupfile.in
echo "-O -i equil1_${i}.mdin -p ../../topology/binder_ejm_31_hmr.parm7 -c binder_heat_${i}.rst7 -o binder_equil1_${i}.mdout -r binder_equil1_${i}.rst7 -x binder_equil1_${i}.nc -ref ../../topology/binder_ejm_31_unrestrained.rst7" >> binder_equil1_groupfile.in
echo "-O -i equil2_${i}.mdin -p ../../topology/binder_ejm_31_hmr.parm7 -c binder_equil1_${i}.rst7 -o binder_equil2_${i}.mdout -r binder_equil2_${i}.rst7 -x binder_equil2_${i}.nc -ref ../../topology/binder_ejm_31_unrestrained.rst7" >> binder_equil2_groupfile.in
done
Now we can run these calculations in parallel using pmemd.cuda.MPI.
mpirun -np 11 pmemd.cuda.MPI -ng 11 -groupfile binder_heat_groupfile.in
mpirun -np 11 pmemd.cuda.MPI -ng 11 -groupfile binder_equil1_groupfile.in
mpirun -np 11 pmemd.cuda.MPI -ng 11 -groupfile binder_equil2_groupfile.in
Hint
Here, note that the usual pmemd commands are included in the group file. On the actual command line, our calls are much simpler now.
Take this for example:
mpirun -np 11 pmemd.cuda.MPI -ng 11 -groupfile binder_heat_groupfile.in
mpirun -np 11: This tells mpirun to use 11 processes (one for each lambda window).
pmemd.cuda.MPI: This is the MPI-enabled version of pmemd.cuda, which allows for parallel execution across multiple processes.
-ng 11: This flag specifies the number of groups (or simulations) to run in parallel, which is 11 in this case.
-groupfile binder_heat_groupfile.in: This specifies the group file that contains the individual pmemd commands for each lambda window. We generated this file in the previous step.
While these won’t take too long to run, they will take longer than the annihilation simulations, as these are full MD simulations (on the order of an hour).
Complex Equilibration
&cntrl
imin = 0
nstlim = 100000
dt = 0.001
irest = 0
ntx = 1
ntxo = 1
ntc = 2
ntf = 1
ntwx = 0
ntwr = 0
ntpr = 125
cut = 10.0
iwrap = 0
ntb = 1
ntp = 0
tempi = 200
temp0 = 298
ntt = 3
gamma_ln = 5.0
barostat = 2
pres0 = 1.01325
taup = 1
rmsd_mask(1) = ':1 & !@H='
rmsd_strength(1)= 0
rmsd_ti(1) = 2
rmsd_type = 0
rmsd_mask(2) = '@CA '
rmsd_strength(2)= 0
rmsd_ti(2) = 3
ig = -1
ifsc = 1
icfe = 1
clambda = REPLACE_WITH_LAMBDA
timask1 = ':1'
timask2 = ''
crgmask = ""
scmask1 = ':1'
scmask2 = ''
scalpha = 0.5
scbeta = 1.0
gti_cut = 1
gti_output = 1
gti_add_sc = 25
gti_scale_beta = 1
gti_cut_sc_on = 8
gti_cut_sc_off = 10
gti_lam_sch = 1
gti_ele_sc = 1
gti_vdw_sc = 1
gti_cut_sc = 2
gti_ele_exp = 2
gti_vdw_exp = 2
gti_syn_mass = 0
gremd_acyc = 1
gti_bat_sc = -1
nmropt = 1
mbar_lambda(1) = 0.00000
mbar_lambda(2) = 0.10000
mbar_lambda(3) = 0.20000
mbar_lambda(4) = 0.30000
mbar_lambda(5) = 0.40000
mbar_lambda(6) = 0.50000
mbar_lambda(7) = 0.60000
mbar_lambda(8) = 0.70000
mbar_lambda(9) = 0.80000
mbar_lambda(10) = 0.90000
mbar_lambda(11) = 1.00000
/
&wt type = 'END',
/
DISANG=rest.in
&wt
TYPE='TEMP0',
ISTEP1=0, ISTEP2=100000,
VALUE1=200, VALUE2=298,
&cntrl
imin = 0
nstlim = 50000
dt = 0.002
irest = 1
ntx = 5
ntxo = 1
ntc = 2
ntf = 1
ntwx = 0
ntwr = 0
ntpr = 125
cut = 10.0
iwrap = 0
ntb = 1
ntp = 0
tempi = 298
temp0 = 298
ntt = 3
gamma_ln = 5.0
barostat = 2
pres0 = 1.01325
taup = 1
rmsd_mask(1) = ':1 & !@H='
rmsd_strength(1)= 0
rmsd_ti(1) = 2
rmsd_type = 0
rmsd_mask(2) = '@CA '
rmsd_strength(2)= 0
rmsd_ti(2) = 3
ig = -1
ifsc = 1
icfe = 1
clambda = REPLACE_WITH_LAMBDA
timask1 = ':1'
timask2 = ''
crgmask = ""
scmask1 = ':1'
scmask2 = ''
scalpha = 0.5
scbeta = 1.0
gti_cut = 1
gti_output = 1
gti_add_sc = 25
gti_scale_beta = 1
gti_cut_sc_on = 8
gti_cut_sc_off = 10
gti_lam_sch = 1
gti_ele_sc = 1
gti_vdw_sc = 1
gti_cut_sc = 2
gti_ele_exp = 2
gti_vdw_exp = 2
gti_syn_mass = 0
gremd_acyc = 1
gti_bat_sc = -1
nmropt = 1
/
&wt type = 'END', /
DISANG=rest.in
&cntrl
imin = 0
nstlim = 250000
dt = 0.004
irest = 1
ntx = 5
ntxo = 1
ntc = 2
ntf = 1
ntwx = 0
ntwr = 0
ntpr = 125
cut = 10.0
iwrap = 0
ntb = 1
ntp = 0
tempi = 298
temp0 = 298
ntt = 3
gamma_ln = 5.0
barostat = 2
pres0 = 1.01325
taup = 1
rmsd_mask(1) = ':1 & !@H='
rmsd_strength(1)= 0
rmsd_ti(1) = 2
rmsd_type = 0
rmsd_mask(2) = '@CA '
rmsd_strength(2)= 0
rmsd_ti(2) = 3
ig = -1
ifsc = 1
icfe = 1
clambda = REPLACE_WITH_LAMBDA
timask1 = ':1'
timask2 = ''
crgmask = ""
scmask1 = ':1'
scmask2 = ''
scalpha = 0.5
scbeta = 1.0
gti_cut = 1
gti_output = 1
gti_add_sc = 25
gti_scale_beta = 1
gti_cut_sc_on = 8
gti_cut_sc_off = 10
gti_lam_sch = 1
gti_ele_sc = 1
gti_vdw_sc = 1
gti_cut_sc = 2
gti_ele_exp = 2
gti_vdw_exp = 2
gti_syn_mass = 0
gremd_acyc = 1
gti_bat_sc = -1
nmropt = 1
/
&wt type = 'END', /
DISANG=rest.in
As with the binder, we will generate the mdins programmatically for each lambda window using a bash for loop.
cd ../complex
# Generate Equilibration MDINs for Each Lambda Window
lambda_values=(0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0)
for i in {0..10}
do
lambda_value=${lambda_values[$i]}
sed "s/REPLACE_WITH_LAMBDA/$lambda_value/g" ../../complex_mdins/heat.mdin > heat_${i}.mdin
sed "s/REPLACE_WITH_LAMBDA/$lambda_value/g" ../../complex_mdins/equil1.mdin > equil1_${i}.mdin
sed "s/REPLACE_WITH_LAMBDA/$lambda_value/g" ../../complex_mdins/equil2.mdin > equil2_${i}.mdin
done
Now, we also need a groupfile for the replica exchange simulations. This file will look a lot like the pmemd.CUDA command calls that we made before, except the groupfile will contain all of the lambda windows to be run in parallel.
The script below will generate the groupfile for the binder equilibration simulations.
rm complex_heat_groupfile.in complex_equil1_groupfile.in complex_equil2_groupfile.in
for i in {0..10}
do
echo "-O -i heat_${i}.mdin -p ../../topology/complex_ejm_31_hmr.parm7 -c annihilate_complex_${i}.rst7 -o complex_heat_${i}.mdout -r complex_heat_${i}.rst7 -x complex_heat_${i}.nc -ref ../../topology/complex_ejm_31_unrestrained.rst7" >> complex_heat_groupfile.in
echo "-O -i equil1_${i}.mdin -p ../../topology/complex_ejm_31_hmr.parm7 -c complex_heat_${i}.rst7 -o complex_equil1_${i}.mdout -r complex_equil1_${i}.rst7 -x complex_equil1_${i}.nc -ref ../../topology/complex_ejm_31_unrestrained.rst7" >> complex_equil1_groupfile.in
echo "-O -i equil2_${i}.mdin -p ../../topology/complex_ejm_31_hmr.parm7 -c complex_equil1_${i}.rst7 -o complex_equil2_${i}.mdout -r complex_equil2_${i}.rst7 -x complex_equil2_${i}.nc -ref ../../topology/complex_ejm_31_unrestrained.rst7" >> complex_equil2_groupfile.in
done
Now we can run these calculations in parallel using pmemd.cuda.MPI.
mpirun -np 11 pmemd.cuda.MPI -ng 11 -groupfile complex_heat_groupfile.in
mpirun -np 11 pmemd.cuda.MPI -ng 11 -groupfile complex_equil1_groupfile.in
mpirun -np 11 pmemd.cuda.MPI -ng 11 -groupfile complex_equil2_groupfile.in
These will take much longer than the binder equilibration simulations, as the complex simulations involve a much larger system size. Expect these to take several hours to complete.
Hint
If you want to run these in the background, you can use nohup or screen to keep the processes running after you log out of your session.
nohup mpirun -np 11 pmemd.cuda.MPI -ng 11 -groupfile complex_heat_groupfile.in &
Note - if you do this, you need to run these commands for each equilibration stage (heat, equil1, equil2) separately, as each command needs to finish before starting the next one.
Running Production Simulations
In the final step, we will run production simulations for each lambda window for both the binder and complex environments. As with the equilibration simulations, we will run a quick equilibration without replica exchange first, followed by the production simulations with replica exchange enabled.
Note
On Generating Independent Trials
In practice, ABFE calculations are often run with multiple independent trials to improve convergence and sampling. This can be achieved by starting each trial from different initial conditions, such as different random seeds or different starting structures. In this tutorial, we will demonstrate how to set up and run a single trial for simplicity. However, in a real-world scenario, you would repeat the equilibration and production steps multiple times with different starting conditions to generate independent trials. This approach helps to ensure that the results are robust and not dependent on a single set of initial conditions.
There are a few ways to achieve this, such as modifying the random seed in the mdin files or using different starting structures for each trial. The key is to ensure that each trial is independent and samples different regions of conformational space.
Binder Production
Like before, we will start with the binder, running each lambda window using the corresponding mdin files provided below.
&cntrl
imin = 0
nstlim = 250000
dt = 0.004
irest = 0
ntx = 1
ntxo = 1
ntc = 2
ntf = 1
ntwx = 0
ntwr = 0
ntpr = 125
cut = 10.0
iwrap = 0
ntb = 1
ntp = 0
tempi = 298
temp0 = 298
ntt = 3
gamma_ln = 5.0
barostat = 2
pres0 = 1.01325
taup = 1
ig = -1
ifsc = 1
icfe = 1
rmsd_mask(1) = ':1 & !@H='
rmsd_strength(1)= 0
rmsd_ti(1) = 2
rmsd_type = 0
clambda = REPLACE_WITH_LAMBDA
timask1 = ':1'
timask2 = ''
crgmask = ""
scmask1 = ':1'
scmask2 = ''
scalpha = 0.5
scbeta = 1.0
gti_cut = 1
gti_output = 1
gti_add_sc = 25
gti_scale_beta = 1
gti_cut_sc_on = 8
gti_cut_sc_off = 10
gti_lam_sch = 1
gti_ele_sc = 1
gti_vdw_sc = 1
gti_cut_sc = 2
gti_ele_exp = 2
gti_vdw_exp = 2
gti_syn_mass = 0
gremd_acyc = 1
/
&cntrl
imin = 0
nstlim = 100
numexchg = 12500
dt = 0.004
irest = 0
ntx = 1
ntxo = 1
ntc = 2
ntf = 1
ntwx = 1250
ntwr = 0
ntpr = 125
cut = 10.0
iwrap = 0
ntb = 1
ntp = 0
tempi = 298
temp0 = 298
ntt = 3
gamma_ln = 5.0
barostat = 2
pres0 = 1.01325
taup = 1
rmsd_mask(1) = ':1 & !@H='
rmsd_strength(1)= 0
rmsd_type = 0
rmsd_ti(1) = 2
ig = -1
ifsc = 1
icfe = 1
ifmbar = 1
bar_intervall = 100
mbar_states = 11
clambda = REPLACE_WITH_LAMBDA
timask1 = ':1'
timask2 = ''
crgmask = ""
scmask1 = ':1'
scmask2 = ''
scalpha = 0.5
scbeta = 1.0
gti_cut = 1
gti_output = 1
gti_add_sc = 25
gti_scale_beta = 1
gti_cut_sc_on = 8
gti_cut_sc_off = 10
gti_lam_sch = 1
gti_ele_sc = 1
gti_vdw_sc = 1
gti_cut_sc = 2
gti_ele_exp = 2
gti_vdw_exp = 2
gti_syn_mass = 0
gremd_acyc = 1
mbar_lambda(1) = 0.00000
mbar_lambda(2) = 0.10000
mbar_lambda(3) = 0.20000
mbar_lambda(4) = 0.30000
mbar_lambda(5) = 0.40000
mbar_lambda(6) = 0.50000
mbar_lambda(7) = 0.60000
mbar_lambda(8) = 0.70000
mbar_lambda(9) = 0.80000
mbar_lambda(10) = 0.90000
mbar_lambda(11) = 1.00000
/
We will generate the mdins programmatically for each lambda window using a bash for loop.
cd ../binder
# Generate Production MDINs for Each Lambda Window
lambda_values=(0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0)
for i in {0..10}
do
lambda_value=${lambda_values[$i]}
sed "s/REPLACE_WITH_LAMBDA/$lambda_value/g" ../../binder_mdins/trial_equil.mdin > trial_equil_lambda_${i}.mdin
sed "s/REPLACE_WITH_LAMBDA/$lambda_value/g" ../../binder_mdins/trial_production.mdin > trial_production_lambda_${i}.mdin
done
We also need a groupfile for the replica exchange simulations, similar to what we did for the equilibration simulations. The script below will generate the groupfile for the binder production simulations.
rm binder_trial_equil_groupfile.in binder_trial_production_groupfile.in
for i in {0..10}
do
echo "-O -i trial_equil_${i}.mdin -p ../../topology/binder_ejm_31_hmr.parm7 -c binder_equil2_${i}.rst7 -o binder_trial_equil_${i}.mdout -r binder_trial_equil_${i}.rst7 -x binder_trial_equil_${i}.nc -ref ../../topology/binder_ejm_31_unrestrained.rst7" >> binder_trial_equil_groupfile.in
echo "-O -i trial_production_${i}.mdin -p ../../topology/binder_ejm_31_hmr.parm7 -c binder_trial_equil_${i}.rst7 -o binder_trial_production_${i}.mdout -r binder_trial_production_${i}.rst7 -x binder_trial_production_${i}.nc -ref ../../topology/binder_ejm_31_unrestrained.rst7" >> binder_trial_production_groupfile.in
done
With that, you are now set up to run production simulations for the binder environment for each lambda window. You can run these simulations in parallel using pmemd.cuda.MPI.
mpirun -np 11 pmemd.cuda.MPI -ng 11 -groupfile binder_trial_equil_groupfile.in
mpirun -np 11 pmemd.cuda.MPI -ng 11 -groupfile binder_trial_production_groupfile.in -rem 3 -remlog remd_binder_ejm31.log
Note
Note the second command includes the flags -rem 3 -remlog remd_binder_ejm31.log. These flags enable replica exchange during the production simulations, allowing for enhanced sampling across the lambda windows. The -rem 3 flag specifies that Hamiltonian molecular dynamics will be used for the replica exchange, while the -remlog flag specifies the name of the log file to record the exchange attempts and outcomes.
With the binder phase calculated, we can now move on to the complex phase.
Complex Production
&cntrl
imin = 0
nstlim = 250000
dt = 0.004
irest = 0
ntx = 1
ntxo = 1
ntc = 2
ntf = 1
ntwx = 0
ntwr = 0
ntpr = 125
cut = 10.0
iwrap = 0
ntb = 1
ntp = 0
tempi = 298
temp0 = 298
ntt = 3
gamma_ln = 5.0
barostat = 2
pres0 = 1.01325
taup = 1
rmsd_mask(1) = ':1 & !@H='
rmsd_strength(1)= 0
rmsd_ti(1) = 2
rmsd_type = 0
rmsd_mask(2) = '@CA '
rmsd_strength(2)= 0
rmsd_ti(2) = 3
ig = -1
ifsc = 1
icfe = 1
clambda = REPLACE_WITH_LAMBDA
timask1 = ':1'
timask2 = ''
crgmask = ""
scmask1 = ':1'
scmask2 = ''
scalpha = 0.5
scbeta = 1.0
gti_cut = 1
gti_output = 1
gti_add_sc = 25
gti_scale_beta = 1
gti_cut_sc_on = 8
gti_cut_sc_off = 10
gti_lam_sch = 1
gti_ele_sc = 1
gti_vdw_sc = 1
gti_cut_sc = 2
gti_ele_exp = 2
gti_vdw_exp = 2
gti_syn_mass = 0
gremd_acyc = 1
gti_bat_sc = -1
nmropt = 1
/
&wt type = 'END', /
DISANG=rest.in
&cntrl
imin = 0
nstlim = 100
numexchg = 12500
dt = 0.004
irest = 0
ntx = 1
ntxo = 1
ntc = 2
ntf = 1
ntwx = 1250
ntwr = 0
ntpr = 125
cut = 10.0
iwrap = 0
ntb = 1
ntp = 0
tempi = 298
temp0 = 298
ntt = 3
gamma_ln = 5.0
barostat = 2
pres0 = 1.01325
taup = 1
ig = -1
rmsd_mask(1) = ':1 & !@H='
rmsd_strength(1)= 0
rmsd_ti(1) = 2
rmsd_type = 0
rmsd_mask(2) = '@CA '
rmsd_strength(2)= 0
rmsd_ti(2) = 3
ifsc = 1
icfe = 1
ifmbar = 1
bar_intervall = 100
mbar_states = 11
clambda = REPLACE_WITH_LAMBDA
timask1 = ':1'
timask2 = ''
crgmask = ""
scmask1 = ':1'
scmask2 = ''
scalpha = 0.5
scbeta = 1.0
gti_cut = 1
gti_output = 1
gti_add_sc = 25
gti_scale_beta = 1
gti_cut_sc_on = 8
gti_cut_sc_off = 10
gti_lam_sch = 1
gti_ele_sc = 1
gti_vdw_sc = 1
gti_cut_sc = 2
gti_ele_exp = 2
gti_vdw_exp = 2
gti_syn_mass = 0
gremd_acyc = 1
gti_bat_sc = -1
nmropt = 1
mbar_lambda(1) = 0.00000
mbar_lambda(2) = 0.10000
mbar_lambda(3) = 0.20000
mbar_lambda(4) = 0.30000
mbar_lambda(5) = 0.40000
mbar_lambda(6) = 0.50000
mbar_lambda(7) = 0.60000
mbar_lambda(8) = 0.70000
mbar_lambda(9) = 0.80000
mbar_lambda(10) = 0.90000
mbar_lambda(11) = 1.00000
/
&wt type = 'END',
/
DISANG=rest.in
We will generate the mdins programmatically for each lambda window using a bash for loop like we did before.
cd ../complex
# Generate Production MDINs for Each Lambda Window
lambda_values=(0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0)
for i in {0..10}
do
lambda_value=${lambda_values[$i]}
sed "s/REPLACE_WITH_LAMBDA/$lambda_value/g" ../../complex_mdins/trial_equil.mdin > trial_equil_lambda_${i}.mdin
sed "s/REPLACE_WITH_LAMBDA/$lambda_value/g" ../../complex_mdins/trial_production.mdin > trial_production_lambda_${i}.mdin
done
We also need a groupfile for the replica exchange simulations, similar to what we did for the equilibration simulations. The script below will generate the groupfile for the complex production simulations.
rm complex_trial_equil_groupfile.in complex_trial_production_groupfile.in
for i in {0..10}
do
echo "-O -i trial_equil_${i}.mdin -p ../../topology/target_ejm_31_hmr.parm7 -c complex_equil2_${i}.rst7 -o complex_trial_equil_${i}.mdout -r complex_trial_equil_${i}.rst7 -x complex_trial_equil_${i}.nc -ref ../../topology/complex_ejm_31_unrestrained.rst7" >> complex_trial_equil_groupfile.in
echo "-O -i trial_production_${i}.mdin -p ../../topology/target_ejm_31_hmr.parm7 -c complex_trial_equil_${i}.rst7 -o complex_trial_production_${i}.mdout -r complex_trial_production_${i}.rst7 -x complex_trial_production_${i}.nc -ref ../../topology/complex_ejm_31_unrestrained.rst7" >> complex_trial_production_groupfile.in
done
With that, you are now set up to run production simulations for both the binder and complex environments for each lambda window. You can run these simulations in parallel using pmemd.cuda.MPI.
mpirun -np 11 pmemd.cuda.MPI -ng 11 -groupfile complex_trial_equil_groupfile.in
mpirun -np 11 pmemd.cuda.MPI -ng 11 -groupfile complex_trial_production_groupfile.in -rem 3 -remlog remd_complex_ejm31.log
Following the completetion of these steps, you will have successfully set up and run absolute binding free energy calculations for the Tyk2 system. You can then proceed to analyze the results and calculate the binding free energies using appropriate analysis tools.
Getting Started
In this tutorial, we will analyze the results of an ABFE calculation completed in the previous steps of this tutorial series. Either use the data generated from those steps, or download the data files here:
Coming Soon!
In this tutorial, there are a few key steps that we will follow:
Extract data from the simulation output files.
Calculate free energy differences using Edgembar.
Create Free Energy HTML Reports
Overview of the Outputs from An ABFE calculation
In the download or in your previous calculations, the production simulations were organized into two main phases: the binder phase and the complex phase.
Each phase consisted of multiple lambda windows, with each window having its own set of output files. For this tutorial, we will focus on analyzing the output log files (.mdout) generated during these calculations, as well as the replica exchange log files (remd_.log).
The mdout files contain detailed information about the energy components and other properties of the system at each step of the simulation. The remd log files provide information about the replica exchange attempts and outcomes, which can be useful for assessing the sampling efficiency across the lambda windows.
Specifically, the mdout files provide output that looks like this:
| TI region 1
NSTEP = 840250 TIME(PS) = 3361.000 TEMP(K) = 294.22 PRESS = 0.0
Etot = -33055.3946 EKtot = 6151.6533 EPtot = -39207.0479
BOND = 0.0000 ANGLE = 0.0000 DIHED = 3.6590
1-4 NB = -0.0696 1-4 EEL = -25.0360 VDWAALS = 6648.6562
EELEC = -45834.2575 EHBOND = 0.0000 RESTRAINT = 0.0000
DV/DL = 79.6779
TEMP0 = 298.0000 REPNUM = 9 EXCHANGE# = 8403
------------------------------------------------------------------------------
Softcore part of the system: 32 atoms, TEMP(K) = 269.57
SC_Etot= 41.8564 SC_EKtot= 22.7663 SC_EPtot = 19.0901
SC_BOND= 6.4851 SC_ANGLE= 11.9679 SC_DIHED = 6.5738
SC_14NB= 0.0000 SC_14EEL= 0.0000 SC_VDW = -5.9367
SC_EEL = 0.0000
SC_RES_DIST= 0.0000 SC_RES_ANG= 0.0000 SC_RES_TORS= 0.0000
SC_EEL_DER= 1.1039 SC_VDW_DER= -15.3796 SC_DERIV = -14.2757
------------------------------------------------------------------------------
| TI region 2
NSTEP = 840250 TIME(PS) = 3361.000 TEMP(K) = 294.32 PRESS = 0.0
Etot = -33078.1609 EKtot = 6128.8870 EPtot = -39207.0479
BOND = 0.0000 ANGLE = 0.0000 DIHED = 3.6590
1-4 NB = -0.0696 1-4 EEL = -25.0360 VDWAALS = 6648.6562
EELEC = -45834.2575 EHBOND = 0.0000 RESTRAINT = 0.0000
DV/DL = 79.6779
TEMP0 = 298.0000 REPNUM = 9 EXCHANGE# = 8403
------------------------------------------------------------------------------
Note that this shows both the energy components, as well as the parts that come from the soft-core region of the system (i.e., the alchemically modified atoms).
Meanwhile, the remd log files provide output that looks like this:
# Replica Exchange log file
# numexchg is 12500
# REMD filenames:
# remlog= remd_complex_ejm31.log
# remtype= rem.type
# Rep#, Neibr#, Temp0, PotE(x_1), PotE(x_2), left_fe, right_fe, Success, Success rate (i,i+1)
...
# exchange 9032
1 2 298.00 -39400.10 -39409.58 -Infinity -1.79 T 0.87
2 1 298.00 -39408.05 -39398.45 1.79 -10.40 T 0.35
3 4 298.00 -39416.77 -39352.95 10.40 -22.46 F 0.06
4 3 298.00 -39332.22 -39390.84 22.45 -33.45 F 0.01
5 6 298.00 -39316.56 -39355.34 32.76 -36.65 F 0.00
6 5 298.00 -39321.48 -39275.80 36.74 -34.91 F 0.00
7 8 298.00 -39129.67 -39121.37 32.64 -25.37 F 0.00
8 7 298.00 -39108.33 -39103.97 23.32 -13.02 F 0.00
9 10 298.00 -39087.24 -39262.19 10.73 -3.20 F 0.07
10 9 298.00 -39262.99 -39081.81 3.20 -0.44 F 0.74
11 -1 298.00 -39197.79 0.00 0.03 0.00 F 0.00
# exchange 9033
1 -1 298.00 -39438.52 0.00 -Infinity -1.79 F 0.87
2 3 298.00 -39467.30 -39324.55 1.79 -10.40 T 0.35
3 2 298.00 -39314.20 -39456.10 10.40 -22.46 T 0.06
4 5 298.00 -39259.84 -39308.14 22.45 -33.45 F 0.01
5 4 298.00 -39282.02 -39223.52 32.76 -36.65 F 0.00
6 7 298.00 -39248.23 -39241.31 36.74 -34.91 F 0.00
7 6 298.00 -39220.57 -39210.90 32.64 -25.37 F 0.00
8 9 298.00 -39378.26 -39145.48 23.32 -13.02 F 0.00
9 8 298.00 -39137.02 -39358.97 10.73 -3.20 F 0.07
10 11 298.00 -39218.70 -39185.63 3.20 -0.44 T 0.74
11 10 298.00 -39185.17 -39218.08 0.03 0.00 T 0.00
Here, the final column is cumulative success rate for exchanges between adjacent replicas, which can be useful for assessing the efficiency of sampling across the lambda windows. Thus, the most important entry in this file is the final success rate for each pair of adjacent replicas.
Danger
It is important to ensure that the exchange success rates between adjacent replicas are reasonable (typically between 20-40%) to ensure adequate sampling across the lambda windows. If the success rates are too low, it may indicate that the lambda spacing is too large or that the system is not equilibrated properly.
To solve low success rates, there are a few strategies that can be employed:
Optimizing the existing lambda schedule to have smaller gaps between windows where success rates are low.
Increasing the simulation time for each window to allow for better equilibration and sampling.
Adding additional intermediate lambda windows to improve overlap between adjacent states. (Often, 24-48 windows are sufficient for ABFE calculations.)
Extracting Data using Amber2Dats
The first step in analyzing the ABFE calculations is to extract the relevant data from the simulation output files. For this, we will use the edgembar-amber2dats tool from the FE-Toolkit suite.
Running Edgembar
Creating Free Energy HTML Reports
Understanding the Results
Running ABFE Calculations with AmberFlow
In this section, we will walk through how to use AmberFlow to set up and run absolute binding free energy calculations (ABFE) for a Tyk2 inhibitor.
The files you will need are located here:
If you have not already done so, please follow the instructions in the amberflow documentation to install amberflow and set up your environment.
Warning
Currently AmberFlow Documentation is not available online - this should be updated when that changes.
Now, we will walk through the steps to get set up to run ABFE calculations with AmberFlow.
The first step is to unpack the tutorial files; here we will unpack them into a directory called amberflow_files:
mkdir amberflow_files
cd amberflow_files
tar -xvzf ../amberflow_files.tar.gz
In this folder, you will have a folder called starting files. This folder contains the starting structure files for the Tyk2 protein and four ligands. The ligands are:
ejm31
ejm42
jmc27
jmc28
As you work through the tutorial, you will be doing certain steps. The pipeline states after each of these steps is shown in blocks below.
Initial Pipeline after Parameterization
Initial Pipeline after Building
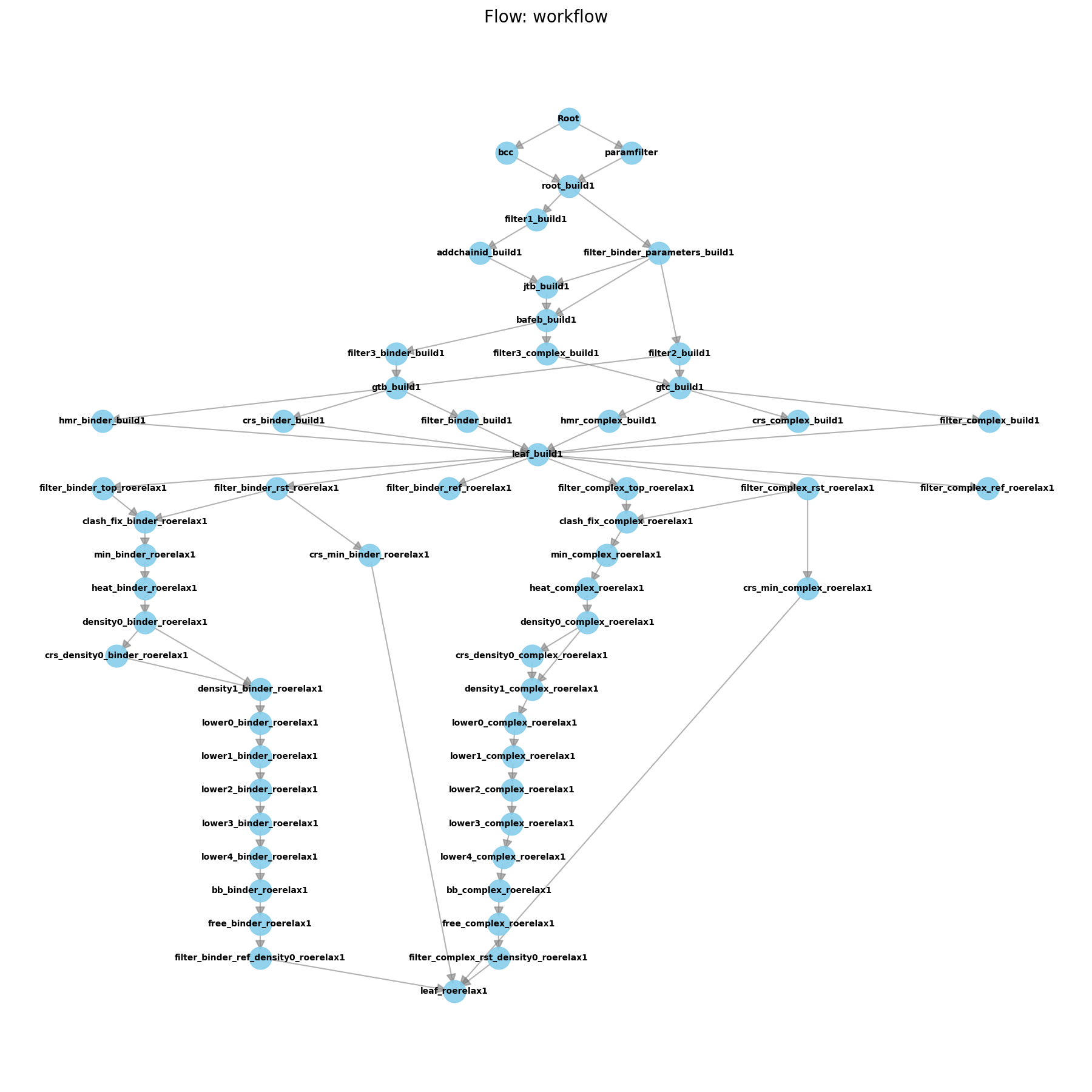
Initial Pipeline after Equilibration
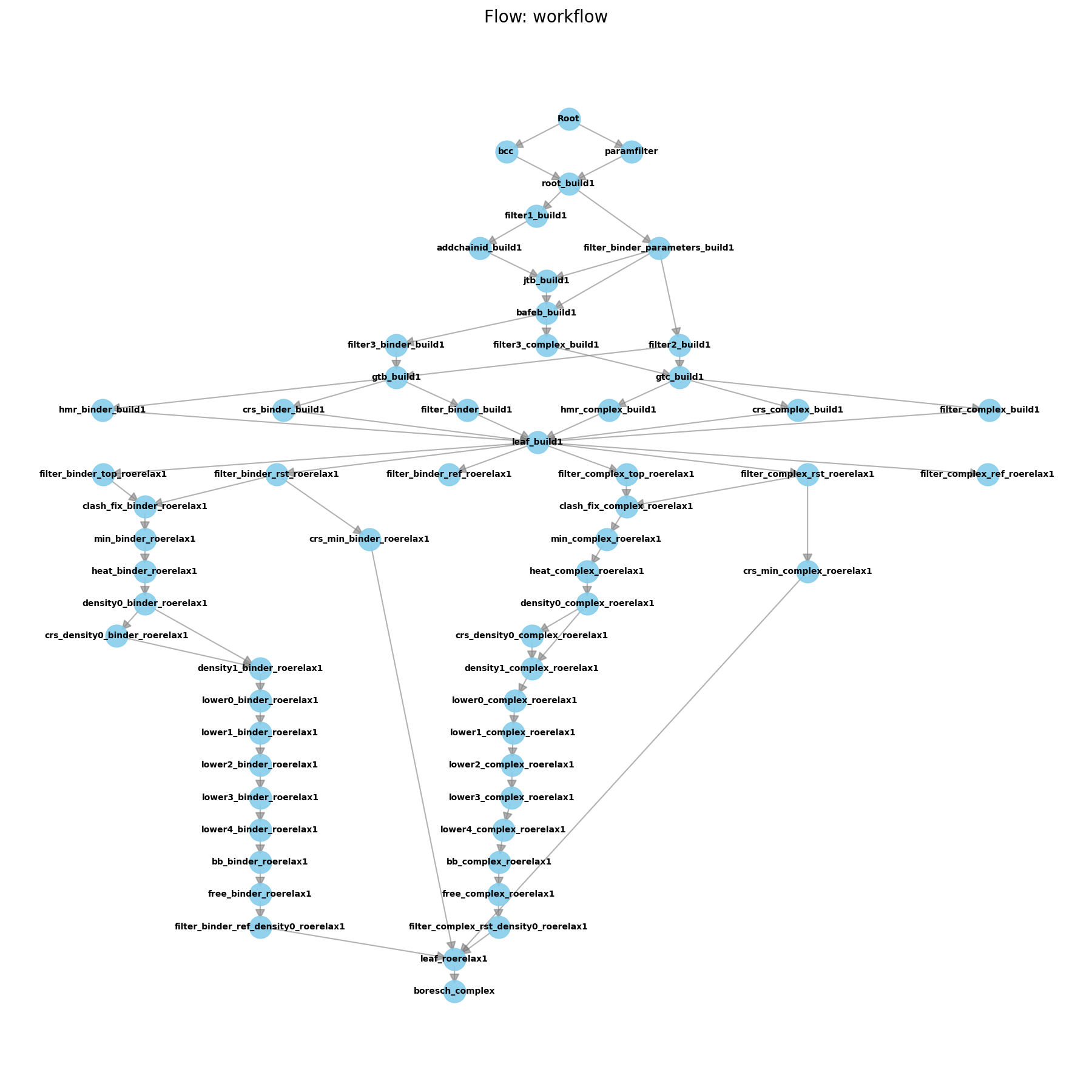
Initial Pipeline after Building

Initial Pipeline after ABFE
Parameterizing the Ligands
For the present tutorial, we will use the GAFF2 forcefield with bcc charges to parameterize the ligands. We will use amberflow’s connection to the ligandparam package for automated ligand parameterization.
To do this, we will start by generating a small amberflow script to run this parameterization. Write an initial python file, e.g. run_parameterization.py, with the following contents:
import logging
from pathlib import Path
from amberflow.schedulers import ReferenceScheduler
from amberflow.worknodes import ParametrizeBinderBCC
from amberflow import Pipeline
home_dir = Path.cwd()
starting_files = home_dir / "tyk2"
# Define the pipeline object
pipa = Pipeline("tyk2_abfe",
cwd=starting_files,
scheduler=ReferenceScheduler(max_cores=10,
max_gpus=1,
allow_gpu_oversubscription=False),
logging_level = logging.DEBUG,
ignore_checkpoint=True)
# Add a worknode for the ligand parameterization.
param_node = ParametrizeBinderBCC(
wnid="bcc",
charge=0,
charge_model='bcc',
skippable=True
)
pipa.append_node(param_node)
pipa.draw()
pipa.launch()
Now this will parameterize the ligands in the tyk2 folder. You can run this script with the following command:
python run_parameterization.py
This will generate output that looks like this:
2025-10-15 11:33:49 - DEBUG - 0.0.1 Loading system jmc28
2025-10-15 11:33:49 - DEBUG - 0.0.1 Loading system ejm42
2025-10-15 11:33:49 - DEBUG - 0.0.1 Loading system jmc27
2025-10-15 11:33:49 - DEBUG - 0.0.1 Loading system ejm31
2025-10-15 11:33:49 - INFO - 0.0.1 Skipping WorkNodeDummy - Root. Status is WorkNodeStatus.COMPLETED.
2025-10-15 11:33:49 - INFO - 0.0.1 Start ParametrizeBinderBCC - bcc
2025-10-15 11:33:59 - INFO - 0.0.1 Node bcc completed on system ejm31
2025-10-15 11:34:06 - INFO - 0.0.1 Node bcc completed on system ejm42
2025-10-15 11:34:09 - INFO - 0.0.1 Node bcc completed on system jmc28
2025-10-15 11:34:10 - INFO - 0.0.1 Node bcc completed on system jmc27
Now, if you look into any of the directories within the tyk2 folder, you will see that there are now new folders present.
The first folder is Root, and is always the base folder created during any amberflow run. If starting with pre-parameterized ligands, you would put those in the base ligand folder and skip the parameterization step. Those files would then be copied into Root, which would be the starting point for that pipeline. In this case, if you look in Root, you will see that it is just a copy of the three pdb files that were in the tyk2 folder to start with.
The second folder is bcc, which contains the output of the ligand parameterization. There are a lot of new files, but the most important are the ones that end with .frcmod, .lib, and .mol2. These are the files that contain the parameters for the ligand. This folder, and the actions that it took is an example of a worknode. Worknodes are the base building blocks of amberflow pipelines. They take in a set of inputs, perform some action, and then generate a set of outputs. Outputs and inputs are referred to as artifacts in amberflow, and can be files (but can also be information such as whether the target is a protein or a nucleic acid). In this case, the binder_pdb files were the input file artifacts, and the frcmod, lib, and mol2 files were the output file artifacts.
If you explore the remaining ligand folders, you will see that they contain the same types of files. Amberflow always runs by applying the same set of worknodes to each system. Other parameterization options exist, such as running a quantum calculation to generate RESP charges, or using the abcg2 charge model. These are all available as different worknodes.
So far, our pipeline looks fairly simple, which you can see in the below figure, but will get more complicated as we move to the next steps.
Building the Systems
The next piece needed for any ABFE calculation is to build the systems. This includes solvating the protein-ligand complex, solvating the ligand alone, and generating the topology files for each of these systems. Such processes involve a number of worknodes, each taking a single step along this process. Here, we will use our first “Flow” to accomplish this task. Within amberflow, a flow is a collection of worknodes that are applied to each system.
To do this, we will copy the parameterization script we used above, and modify it to include a flow for building the systems. By leaving skippable=True in the parameterization worknode, we won’t redo the work if it has already been done, but will instead just automatically use the existing output artifacts.
For completeness, here is the full script, which you can save as run_abfe_build.py:
# OLD CODE
import logging
from pathlib import Path
from amberflow.schedulers import ReferenceScheduler
from amberflow.worknodes import ParametrizeBinderBCC
from amberflow import Pipeline
home_dir = Path.cwd()
starting_files = home_dir / "tyk2"
# Define the pipeline object
pipa = Pipeline("tyk2_abfe",
cwd=starting_files,
scheduler=ReferenceScheduler(max_cores=10,
max_gpus=1,
allow_gpu_oversubscription=False),
logging_level = logging.DEBUG,
ignore_checkpoint=True)
# Add a worknode for the ligand parameterization.
param_node = ParametrizeBinderBCC(
wnid="bcc",
charge=0,
charge_model='bcc',
skippable=True
)
pipa.append_node(param_node)
# NEW CODE
from amberflow.worknodes import Filter
from amberflow.artifacts import BinderLigandPDB
filter1 = Filter(
wnid=f"paramfilter",
artifact_types=(BinderLigandPDB,),
skippable=True,
fail_if_no_artifacts=True,
single=True,
in_or_out="out"
)
from amberflow.flows import FlowBuild1
# Write a flow to build the systems
build_flow = FlowBuild1(
ffpopt=False,
aq_buffer=16.0,
com_buffer=10.0
)
# append the build flow to the pipeline
pipa.append_node(filter1, pipa.root, update_leaves="none")
pipa.append_flow(build_flow)
pipa.add_edge(filter1, "root_build1")
# OLD CODE
pipa.draw()
pipa.launch()
Now here, we have appended two things to the pipeline. First, we appended a filter worknode. This worknode is used to filter the artifacts that are passed between worknodes. In this case, we are filtering so that the binder_pdb in the bcc worknode from the parameterization is used over the binder pdb in the root folder. Filters are important when you have multiple worknodes that generate the same type of artifact, and you want to make sure that the correct one is passed to the next worknode. While in general the most recent artifact is the one that should be used, there are cases where this is not true, and so filters allow (and amberflow requires) that you explicitly pass only the artifacts you want to the next worknode.
Warning
If you do not use a filter in this case, amberflow will raise an error and not run the pipeline. To see this behavior, you can try commenting out the filter lines and running the script again. You will see an error message indicating that there are multiple artifacts of the same type and that a filter is needed to resolve the ambiguity.
The second thing we appended was a flow, which is a collection of worknodes that are applied in sequence. You can see that the flow diagram generated by the pipa.draw() command has grown significantly. The FlowBuild1 flow contains all the worknodes needed to build the solvated protein-ligand and ligand-only systems, as well as generate the topology files for each of these systems. You’ll also see that there is quite a bit of branching in the flow. Worknodes in the pipeline happen from top to bottom, so any worknode that is below another worknode will not be run until the worknode above it has completed.
Now, if you take a look in any of the ligand folders you will see that there are now MANY new folders present. Each of these folders corresponds to a worknode in the flow. The names of the folders correspond to the wnid parameter that was used when creating the workdnode (with flows having certain defaults for wnid). The first folder in the flow will have “root” in the name (in this case, we used this above when we called “root_build1” in the add_edge command). This is the starting point for the flow, and will contain a copy of all the files that were present in the folder before the flow was run. Then, the final folder of each flow will be the leaf folder, which contains the final output files of the flow. In this case, the leaf folders are called “leaf_build1”.
If you look at this leaf folder for the ligand ejm31, you will see that it contains the following files:
binder_ejm31.parm7: The topology file for the ligand-only system.
binder_ejm31.rst7: The restart file for the ligand-only system.
binderref_ejm31.rst7: The restart file for the ligand-only system that will be used as a reference for restraints.
complex_ejm31.parm7: The topology file for the protein-ligand complex.
complex_ejm31.rst7: The restart file for the protein-ligand complex.
complexref_ejm31.rst7: The restart file for the protein-ligand complex that will be used as a reference for restraints.
If you visualize these structures for each ligand, you will see that they are all built and solvated with the same number of water molecules, and that a physiological concentration of NaCl ions have been added to each system.
Running End State Equilibrations
Warning
The remainder of this tutorial requires running real amber calculations which can be time consuming, and maybe not compatible with local resources depending on your current environment. If you want to run these calculations on a different system, copy the entire directory structure that you have run thus far to that system and then run the next steps there. Amberflow, Amber, and Fe-Toolkit must be installed on that system.
Note
When running on another system, you may want to modify the scheduler parameters in the pipeline definition to match the resources available on that system. Currently, this script uses ReferenceScheduler which runs 1 system at a time, but on systems with multiple GPUs, you may want to use DefaultScheduler instead (and then modify max_cores and max_gpus accordingly). Also, usually allow_gpu_oversubscription should be set to True on systems with multiple GPUs as this will better use the available resources.
from amberflow.schedulers import DefaultScheduler
pipa = Pipeline("tyk2_abfe",
cwd=starting_files,
scheduler=DefaultScheduler(max_cores=40,
max_gpus=4,
allow_gpu_oversubscription=True),
logging_level = logging.DEBUG,
ignore_checkpoint=True)
The next step in the ABFE calculation is to equilibrate the systems. Roughly, this involes a number of minimization, heating, and equilibration steps. AmberFlow provides a flow, FlowRoeRelax1, that contains all the worknodes needed to perform these steps:
Minimize the system with restraints on the protein + ligand
Minimize the system without restraints
Fix Clashes
Heat the system from 0K to 300K with restraints on the protein + ligand
Equilibrate the density in a series of short simulations with restraints on the protein + ligand
Equilibrate the system at 300K and 1 atm with restraints on the protein
Slowly reduce the restraints on the protein and ligand
Equilibrate the system at 300K and 1 atm without restraints
To add this flow to the pipeline, we will copy the script from above and modify it to include this new flow. Save this as run_abfe_equil.py:
# OLD CODE
import logging
from pathlib import Path
from amberflow.schedulers import ReferenceScheduler
from amberflow.worknodes import ParametrizeBinderBCC
from amberflow import Pipeline
home_dir = Path.cwd()
starting_files = home_dir / "tyk2"
# Define the pipeline object
pipa = Pipeline("tyk2_abfe",
cwd=starting_files,
scheduler=ReferenceScheduler(max_cores=10,
max_gpus=1,
allow_gpu_oversubscription=False),
logging_level = logging.DEBUG,
ignore_checkpoint=True)
# Add a worknode for the ligand parameterization.
param_node = ParametrizeBinderBCC(
wnid="bcc",
charge=0,
charge_model='bcc',
skippable=True
)
pipa.append_node(param_node)
from amberflow.worknodes import Filter
from amberflow.artifacts import BinderLigandPDB
filter1 = Filter(
wnid=f"paramfilter",
artifact_types=(BinderLigandPDB,),
skippable=True,
fail_if_no_artifacts=True,
single=True,
in_or_out="out"
)
from amberflow.flows import FlowBuild1
# Write a flow to build the systems
build_flow = FlowBuild1(
ffpopt=False,
aq_buffer=16.0,
com_buffer=10.0
)
# append the build flow to the pipeline
pipa.append_node(filter1, pipa.root, update_leaves="none")
pipa.append_flow(build_flow)
pipa.add_edge(filter1, "root_build1")
# New Code
from amberflow.flows import FlowRoeRelax1
relax_flow = FlowRoeRelax1(
complex_full_restraint_mask=":1-289",
complex_noh_restraint_mask=":1-289&!@H=",
binder_full_restraint_mask=":1",
binder_noh_restraint_mask=":1&!@H=",
strong_restraint_wt=50,
bb_restraint_mask="@CA,N,C | :1&!@H=",
bb_restraint_wt=5,
complex_free_restraint_mask=None,
complex_free_restraint_wt=5,
nstlim=1000000,
nstlim_density_equilibration=20000,
skippable=True,
)
pipa.append_flow(relax_flow)
# OLD CODE
pipa.draw()
pipa.launch()
Here, we have appended the FlowRoeRelax1 flow to the pipeline. This flow contains all the worknodes needed to perform the equilibration steps outlined above. Note here that we have specified a number of parameters for the flow, such as the restraint masks and weights. These parameters are important for ensuring that the equilibration is performed correctly. You will need to modify these parameters based on your system. The parameters used here are appropriate for the Tyk2 system, which has 288 residues and a single ligand residue (hence 1-289) in the complex restraint mask (which includes the ligand as residue 1).
Now if you run this script, you will see that the flow diagram has grown even more, with the new flow connecting to the leaf folder of the previous flow. This is because the output artifacts of the previous flow are the input artifacts for this flow. The leaf folder of this new flow is called “leaf_roerelax1”, and contains the final equilibrated structures for each system.
If you look in this folder for the ligand ejm31, you will see that it contains the following files:
binder_ejm31.mdout: The amber mdout file for the ligand-only system.
binder_ejm31.parm7: The topology file for the ligand-only system.
binder_ejm31.rst7: The restart file for the ligand-only system.
binderref_ejm31.rst7: The restart file for the ligand-only system that will be used as a reference for restraints.
binder_ejm31.nc: The amber trajectory file for the ligand-only system.
complex_ejm31.mdout: The amber mdout file for the protein-ligand complex.
complex_ejm31.parm7: The topology file for the protein-ligand complex.
complex_ejm31.rst7: The restart file for the protein-ligand complex.
complexref_ejm31.rst7: The restart file for the protein-ligand complex that will be used as a reference for restraints.
complex_ejm31.nc: The amber trajectory file for the protein-ligand complex.
node.log: The amberflow log file for the flow.
Note - these are all the files that would be necessary to run ABFE calculations. However, we are not quite ready to run ABFE yet. There is one more piece we need to add to the pipeline before we can run ABFE calculations, which is to determine the Boresch restraints that will be used to restrain the ligand in the binding site during the ABFE calculations. This will be covered in the next section.
Determining Boresch Restraints Using AmberFlow
Now we are almost ready for ABFE! The last piece we need before running actual ABFE calculatioons is to determine the Boresch restraints that will be used to restrain the ligand in the binding site when its interactions are decoupled from the protein during Hamiltonian replica exchange simulations. As you might surmise by this point, AmberFlow has a WorkNode that can do this for us!
# OLD CODE
import logging
from pathlib import Path
from amberflow.schedulers import ReferenceScheduler
from amberflow.worknodes import ParametrizeBinderBCC
from amberflow import Pipeline
home_dir = Path.cwd()
starting_files = home_dir / "tyk2"
# Define the pipeline object
pipa = Pipeline("tyk2_abfe",
cwd=starting_files,
scheduler=ReferenceScheduler(max_cores=10,
max_gpus=1,
allow_gpu_oversubscription=False),
logging_level = logging.DEBUG,
ignore_checkpoint=True)
# Add a worknode for the ligand parameterization.
param_node = ParametrizeBinderBCC(
wnid="bcc",
charge=0,
charge_model='bcc',
skippable=True
)
pipa.append_node(param_node)
from amberflow.worknodes import Filter
from amberflow.artifacts import BinderLigandPDB
filter1 = Filter(
wnid=f"paramfilter",
artifact_types=(BinderLigandPDB,),
skippable=True,
fail_if_no_artifacts=True,
single=True,
in_or_out="out"
)
from amberflow.flows import FlowBuild1
# Write a flow to build the systems
build_flow = FlowBuild1(
ffpopt=False,
aq_buffer=16.0,
com_buffer=10.0
)
# append the build flow to the pipeline
pipa.append_node(filter1, pipa.root, update_leaves="none")
pipa.append_flow(build_flow)
pipa.add_edge(filter1, "root_build1")
from amberflow.flows import FlowRoeRelax1
relax_flow = FlowRoeRelax1(
complex_full_restraint_mask=":1-289",
complex_noh_restraint_mask=":1-289&!@H=",
binder_full_restraint_mask=":1",
binder_noh_restraint_mask=":1&!@H=",
strong_restraint_wt=50,
bb_restraint_mask="@CA,N,C | :1&!@H=",
bb_restraint_wt=5,
complex_free_restraint_mask=None,
complex_free_restraint_wt=5,
nstlim=1000000,
nstlim_density_equilibration=20000,
skippable=True,
)
pipa.append_flow(relax_flow)
# New Code
from amberflow.worknodes import GetBoresch
boresch_complex = GetBoresch(wnid="boresch_complex", skippable=True)
pipa.append_node(boresch_complex, update_leaves="add")
# OLD CODE
pipa.draw()
pipa.launch()
This worknode will analyze the equilibrated complex structure and determine the optimal set of Boresch restraints to use for ABFE calculations. These restraints will then be used in the ABFE calculations to restrain the ligand in the binding site. If you look into the boresch_complex folder within ejm_31, you will see the output of this command after running it.
The file produced looks like this:
&rst iat=1489,15,0
r1=0.00000,r2=4.46147,r3=4.46147,r4=999.000,rk2=14.62, rk3=14.62/
&rst iat=1477,1489,15,0
r1=-180.00000,r2=84.48356,r3=84.48356,r4=180.000,rk2=54.14, rk3=54.14/
&rst iat=1489,15,14,0
r1=-180.00000,r2=66.83900,r3=66.83900,r4=180.000,rk2=37.54, rk3=37.54/
&rst iat=1490,1477,1489,15,0
r1=-180.00000,r2=14.62579,r3=14.62579,r4=180.000,rk2=42.50, rk3=42.50/
&rst iat=1477,1489,15,14,0
r1=-180.00000,r2=28.80971,r3=28.80971,r4=180.000,rk2=64.34, rk3=64.34/
&rst iat=1489,15,14,16,0
r1=-180.00000,r2=-138.45151,r3=-138.45151,r4=180.000,rk2=52.87, rk3=52.87/
Running the ABFE Flow within AmberFlow
Note
This next section deals with running ABFE calculations. ABFE calculations are compute expensive, and you may want to run this on supercomputing resources if you have access to them. These runs may take up to 48 hours per ligand on a single GPU, so plan accordingly. If you do not have access to supercomputing resources, you can still run this tutorial but you may want to run far fewer steps and reducing the number of lambda windows. Such changes will make your results innacurate, but will allow you to see how the pipeline works without needing to wait for days.
In this section of the tutorial, we will add the final flow to the pipeline, which is the FlowABFE7 flow. This flow contains all the worknodes needed to perform ABFE calculations using Hamiltonian replica exchange with Alchemically Enhanced Sampling. This flow will use the equilibrated structures from the previous flow, as well as the Boresch restraints determined in the previous section.
The rough steps involved in this flow are:
Annihilated the ligand in the complex and ligand-only systems using a series of lambda windows.
Reinitialize and Reheat the system from 100 K to 300 K
Equilibrate the Lambda Windows
Run Production Hamiltonian Replica Exchange Simulations
Analyze the results using FE-TOOLKIT and EDGEMBAR to determine the free energy of binding.
To add this flow to the pipeline, we will copy the script from above and modify it to include this new flow. Save this as run_abfe.py:
# OLD CODE
import logging
from pathlib import Path
from amberflow.schedulers import ReferenceScheduler
from amberflow.worknodes import ParametrizeBinderBCC
from amberflow import Pipeline
home_dir = Path.cwd()
starting_files = home_dir / "tyk2"
# Define the pipeline object
pipa = Pipeline("tyk2_abfe",
cwd=starting_files,
scheduler=ReferenceScheduler(max_cores=10,
max_gpus=1,
allow_gpu_oversubscription=False),
logging_level = logging.DEBUG,
ignore_checkpoint=True)
# Add a worknode for the ligand parameterization.
param_node = ParametrizeBinderBCC(
wnid="bcc",
charge=0,
charge_model='bcc',
skippable=True
)
pipa.append_node(param_node)
from amberflow.worknodes import Filter
from amberflow.artifacts import BinderLigandPDB
filter1 = Filter(
wnid=f"paramfilter",
artifact_types=(BinderLigandPDB,),
skippable=True,
fail_if_no_artifacts=True,
single=True,
in_or_out="out"
)
from amberflow.flows import FlowBuild1
# Write a flow to build the systems
build_flow = FlowBuild1(
ffpopt=False,
aq_buffer=16.0,
com_buffer=10.0
)
# append the build flow to the pipeline
pipa.append_node(filter1, pipa.root, update_leaves="none")
pipa.append_flow(build_flow)
pipa.add_edge(filter1, "root_build1")
from amberflow.flows import FlowRoeRelax1
relax_flow = FlowRoeRelax1(
complex_full_restraint_mask=":1-289",
complex_noh_restraint_mask=":1-289&!@H=",
binder_full_restraint_mask=":1",
binder_noh_restraint_mask=":1&!@H=",
strong_restraint_wt=50,
bb_restraint_mask="@CA,N,C | :1&!@H=",
bb_restraint_wt=5,
complex_free_restraint_mask=None,
complex_free_restraint_wt=5,
nstlim=1000000,
nstlim_density_equilibration=20000,
skippable=True,
)
pipa.append_flow(relax_flow)
from amberflow.worknodes import GetBoresch
boresch_complex = GetBoresch(wnid="boresch_complex", skippable=True)
pipa.append_node(boresch_complex, update_leaves="add")
# New Code
from amberflow.flows import FlowABFE7
binder_windows = [
0.00000000, 0.13098000, 0.21201000, 0.26401000, 0.30636000, 0.34433000,
0.37938000, 0.41236000, 0.44434000, 0.47551000, 0.50621000, 0.53668000,
0.56698000, 0.59718000, 0.62685000, 0.65568000, 0.68242000, 0.70731000,
0.73179000, 0.75770000, 0.78666000, 0.82089000, 0.86930000, 1.00000000,
]
complex_windows = [
0.00000000, 0.13102000, 0.21119000, 0.26280000, 0.30471000, 0.34202000,
0.37647000, 0.40898000, 0.44048000, 0.47140000, 0.50177000, 0.53204000,
0.56183000, 0.59113000, 0.62058000, 0.65039000, 0.68020000, 0.70877000,
0.73602000, 0.76324000, 0.79211000, 0.82503000, 0.86947000, 1.00000000,
]
abfe_flow = FlowABFE7(
binder_windows=binder_windows,
complex_windows=complex_windows,
heating_restraint_mask=":1-289&!@H=",
heating_restraint_wt=5,
complex_rmsd_mask1=":1 & !@H=",
complex_rmsd_strength1_heat=0,
complex_rmsd_strength1_equil1=0,
complex_rmsd_strength1_equil2=0,
complex_rmsd_strength1_trial_equil=0,
complex_rmsd_strength1_trial=0,
complex_rmsd_ti1=2,
complex_rmsd_mask2="@CA ",
complex_rmsd_strength2_heat=0,
complex_rmsd_strength2_equil1=0,
complex_rmsd_strength2_equil2=0,
complex_rmsd_strength2_trial_equil=0,
complex_rmsd_strength2_trial=0,
complex_rmsd_ti2=3,
binder_rmsd_mask=":1 & !@H=",
binder_rmsd_strength_heat=0,
binder_rmsd_strength_equil1=0,
binder_rmsd_strength_equil2=0,
binder_rmsd_strength_trial_equil=0,
binder_rmsd_strength_trial=0,
binder_rmsd_ti=2,
complex_rmsd_type=0,
binder_rmsd_type=0,
nstlim_heat=100000,
nstlim_equil1=50000,
nstlim_equil2=250000,
nstlim_trial_equil=250000,
nstlim_trial=100,
bar_intervall=100,
ntwx_trial=1250,
numexchg=12500,
nmropt=True,
start_trial=1,
end_trial=1,
skippable=True,
skip_analysis=False,
max_systems_binder = 1,
max_systems_complex = 1,
)
pipa.append_flow(abfe_flow)
# OLD CODE
pipa.draw()
pipa.launch()
Note that in the above script, we have specified a wide-range of parameters for the ABFE calculations, most of which are left at their default values. Many of these are related to features that we won’t be using for this tutorial, such as RMSD and lambda-dependent RMSD restraints (set to 0 above); however, many of the other parameters have significant impacts on calculated results.
In general, anything marked nstlim_XXX is the number of steps to run for that phase of the calculation. The values used above are appropriate for a production ABFE calculation, but you may want to reduce these values if you are running on a local machine and want to see results quickly. The total production length in steps is determined by bar_intervall*numexchg.
start_trial and end_trial determine how many trials to run. Each trial is an independent repeat of an equilibration and production run. More trials will give better statistics, but will also take longer to run. For a production calculation, you will want to run at least 3 trials, but for testing purposes, you can set both of these to 1 to just run a single trial.
max_systems_binder and max_systems_complex determine how many systems to run in parallel. This is useful if you have multiple GPUs available, as you can run multiple systems at once to better utilize the available resources. However, if you are running on a single GPU, you will want to set these to 1 to avoid oversubscribing the GPU.
As you can see from the flow diagram, the pipeline is quite large!
Analyzing the Results
Now, the experimental binding free energies for these ligands are known. You can compare your calculated results to the experimental values to see how well the calculations performed. The experimental values are as follows:
jcm27 : -11.28 kcal/mol
jcm28 : -10.98 kcal/mol
ejm31 : -9.56 kcal/mol
ejm42 : -9.78 kcal/mol
After running the ABFE flow, you will see that in each ligand folder there is now a folder called “leaf_abfe7”. This folder contains the summary output files for the ABFE calculations. There are two main files of interest:
fe_data.csv: This file contains the free energy values summarized for all four ligands (if you ran them as a single flow). this file contains the calculated binding free energy for each ligand, as well as the associated error estimates.
edge_{ligand}.html: This file contains a detailed analysis of the free energy calculations for each ligand. It includes plot of the du-dlambda profile, as well as other useful information about the calculations.
Note
Each ligands html file is located in that ligand’s leaf_abfe7 folder, whereas, the fe_data.csv file contains information about all ligands.
For more information on the html files for analysis, please check out the relevant tutorial.